Toward universal cell embeddings: integrating single-cell RNA-seq datasets across species with SATURN
SATURN is a deep learning method for learning universal cell embeddings that encodes genes’ biological properties using protein language models. SATURN integrates datasets profiled from different species regardless of their genomic similarity and can detect functionally related genes coexpressed across species.
Cell mapping consortia efforts have generated large-scale single-cell datasets comprising hundreds of thousands of cells with the goal of uncovering underlying cellular processes. However, current analyses remain limited in their ability to jointly analyze datasets generated across different species. Such joint analysis offers great potential for understanding fundamental evolutionary processes such as identifying cell types that are conserved across species and identifying the corresponding gene programs that drive similarities and differences of such cell types.
We develop SATURN (Species Alignment Through Unification of Rna and proteiNs), a deep learning approach that integrates cross-species single-cell RNA-sequencing (scRNA-seq) datasets by coupling gene expression with protein embeddings generated by large protein language models. SATURN is uniquely able to perform multispecies differential expression analysis revealing functionally related groups of genes coexpressed across species. By mapping single-cell datasets generated with different genes to a joint embedding space, SATURN takes important steps toward universal cell embeddings.
Publication
Toward universal cell embeddings: integrating single-cell RNA-seq datasets across species with SATURN.
Yanay Rosen*, Maria Brbic*, Yusuf Roohani*, Kyle Swanson, Ziang Li, Jure Leskovec.
Nature Methoods, 2024.
@article{saturn2024,
title={Toward universal cell embeddings: integrating single-cell RNA-seq datasets across species with SATURN},
author={Rosen, Yanay and Brbi’c, Maria and Roohani, Yusuf and Swanson, Kyle and Li, Ziang
and Leskovec, Jure},
journal={Nature Methods},
year={2024},
}
Overview of SATURN
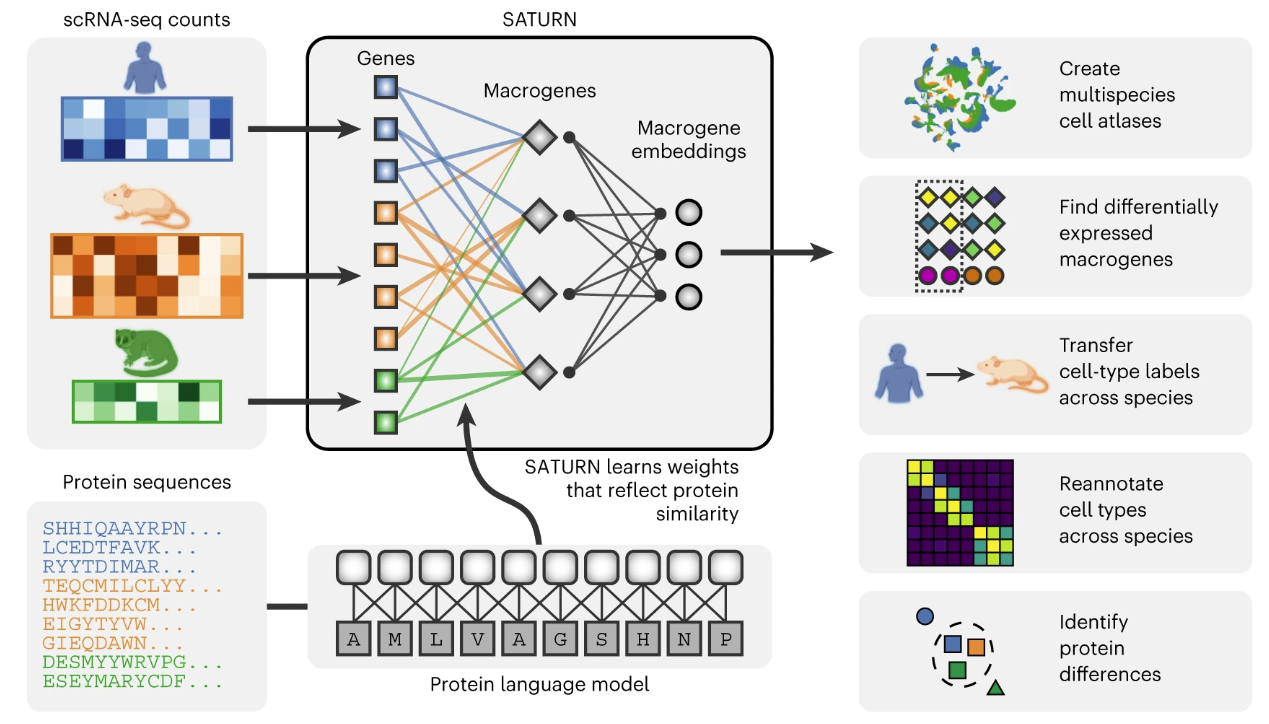
SATURN integrates scRNA-seq datasets generated from different species with different genes by mapping them to a joint low-dimensional embedding space using gene expression and protein representations. SATURN takes as input: (i) scRNA-seq count data from one or multiple species, (ii) protein embeddings generated by a large protein embedding language model, and (iii) initial within-species cell annotations
Given gene expression and protein embeddings, SATURN learns an interpretable feature space shared between multiple species. We refer to this space as a macrogene space and it represents a joint space composed of genes inferred to be functionally related based on the similarity of their protein embeddings. The importance of a gene to a macrogene is defined by a neural network weight—the stronger the importance, the higher the value of the weight that connects the gene to the macrogene.
Case study: Creating multispecies cell atlases
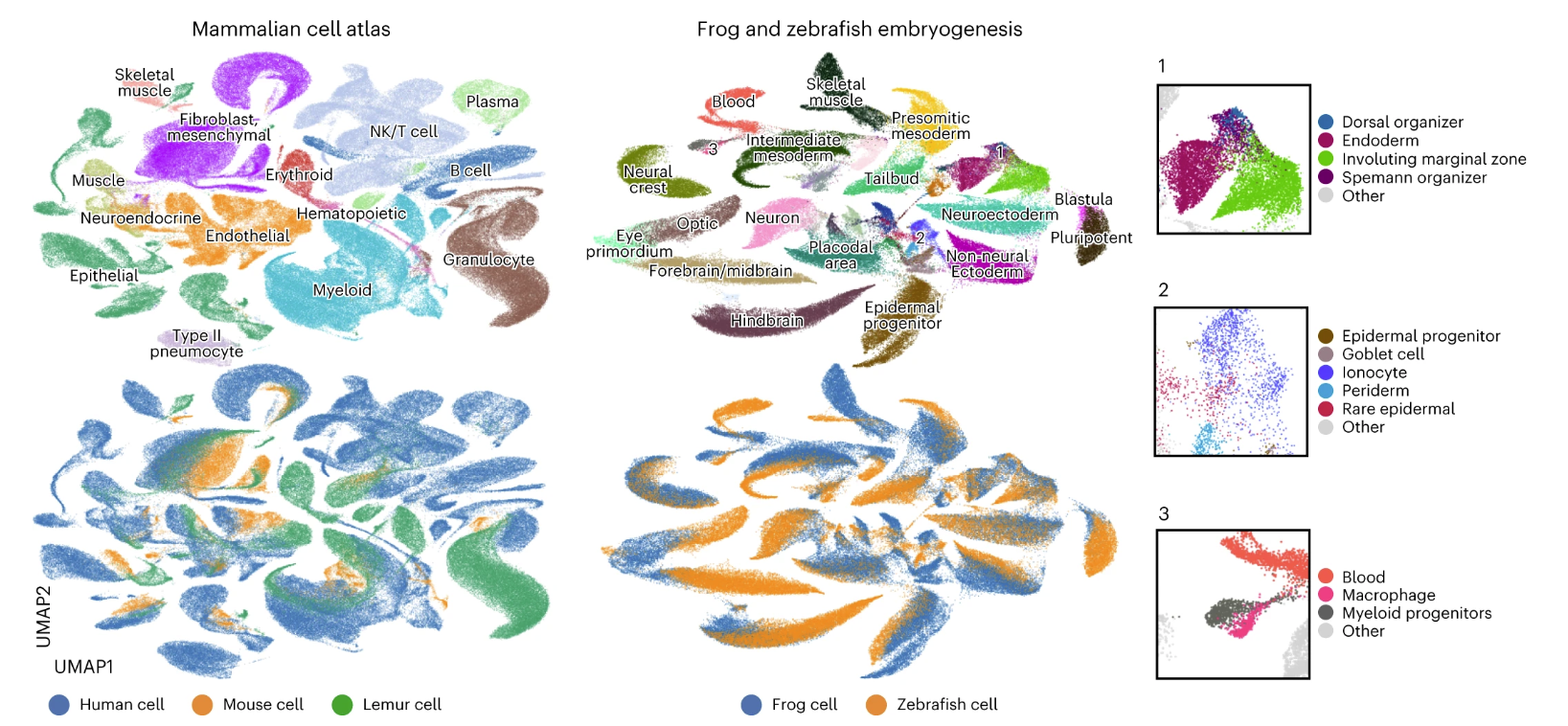
We applied SATURN to integrate large-scale single-cell atlas datasets generated from human (Tabula Sapiens), mouse lemur (Tabula Microcebus) and mouse (Tabula Muris), creating the mammalian cell atlas of 335,000 cell. We found that major cell types aligned well across three species such as T cells, B cells and muscle cells, and then we analyzed the alignment on a per-tissue level. For example, in muscle, we found a small subcluster of cells labeled as mouse macrophages that grouped with human and lemur granulocytes, while the rest of cells labeled as mouse macrophages aligned with human and lemur macrophages.
We next applied SATURN to a multispecies dataset of frog (97,000 cells) and zebrafish (63,000 cells) embryogenesis. SATURN aligned evolutionarily related cell types between these two remote species . We inspected small clusters that are aligned by SATURN, but their ground-truth cell-type annotations differ. We find that these clusters indeed correspond to related cell types. For example, SATURN integrated zebrafish early-stage macrophages and frog myeloid progenitors, which can differentiate into macrophages.
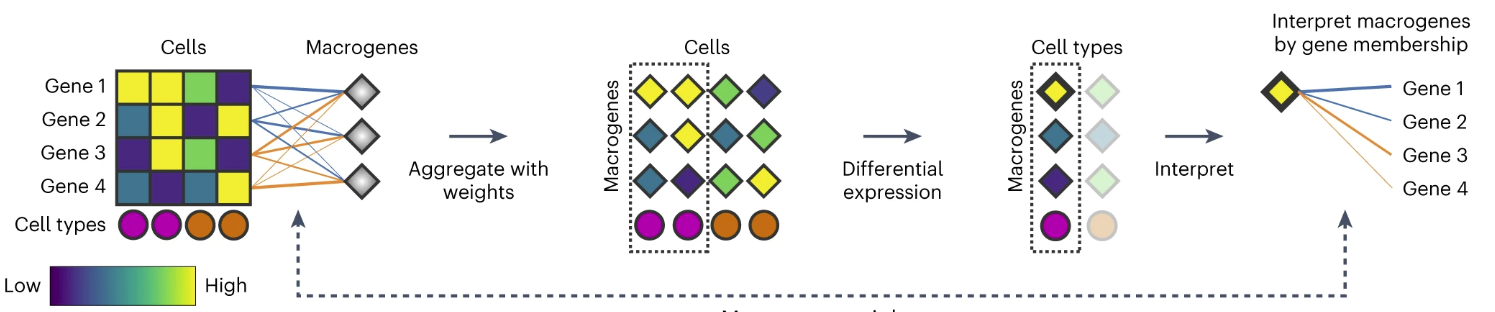
SATURN performs differential expression on macrogenes
SATURN extends differential expression analysis to a multispecies setting. Instead of performing differential expression analysis on individual genes, which is highly limited when datasets do not share genes, SATURN performs differential expression on macrogenes, which enables characterization of cell-type-specific macrogenes across different datasets. To perform differential expression on macrogenes, SATURN first aggregates the contributions of individual genes to macrogenes using gene–macrogene neural network weights.
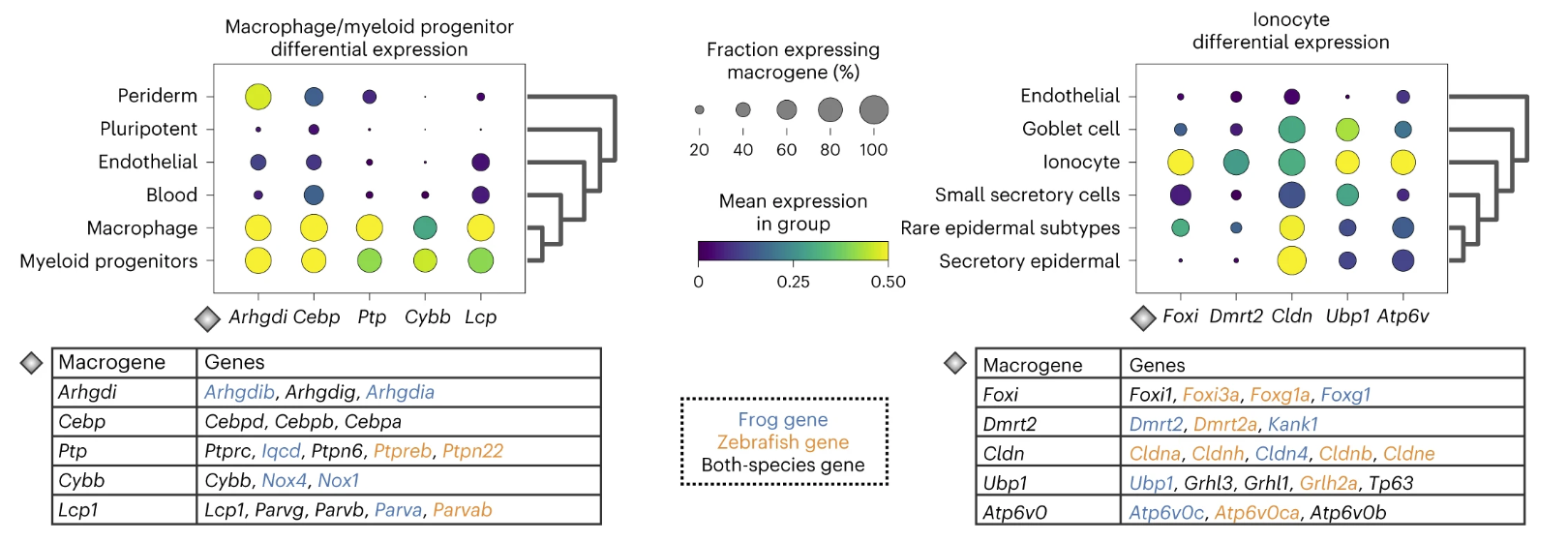
We conduct macrogene differential expression analysis in frog and zebrafish embryogenesis datasets. We demonstrate examples for the macrophage/myeloid progenitor cluster and for the ionocytes cluster. In particular, we show the top five differentially expressed macrogenes and their corresponding highly weighted genes that characterize them, and we name each macrogene according to the gene with the highest weight to that macrogene.
Code
A PyTorch implementation of SATURN is available on GitHub.
Contributors
The following people contributed to this work:
Yanay Rosen*
Maria Brbic*
Yusuf Roohani*
Kyle Swanson
Ziang Li
Jure Leskovec