Concept Learners for Few-Shot Learning
COMET is an inherently interpretable meta-learning method that learns generalizable representations along human-understandable concept dimensions.
Developing algorithms that are able to generalize to a novel task given only a few labeled examples represents a fundamental challenge in closing the gap between machine- and human-level performance. The core of human cognition lies in the structured, reusable concepts that help us to rapidly adapt to new tasks and provide reasoning behind our decisions. Motivated by the human cognition, we advocate that the lack of structure is the missing piece in improving generalization ability of the current meta-learners.
Here we develop a meta-learning method, called COMET, that learns generalizable representations along human-interpretable concept dimensions. COMET learns mappings of high-level concepts into semi-structured metric spaces, and effectively combines the outputs of independent concept learners. Three key aspects lead to a strong generalization ability: (i) semi-structured representation learning, (ii) concept-specific metric spaces described with concept prototypes, and (iii) ensembling of many models.
Publication
Concept Learners for Few-Shot Learning.
Kaidi Cao*, Maria Brbić*, Jure Leskovec.
ICLR, 2021.
@inproceedings{comet21,
title={Concept Learners for Few-Shot Learning},
author={Cao, Kaidi and Brbi\’c, Maria and Leskovec, Jure},
booktitle={International Conference on Learning Representations (ICLR)},
year={2021},
}
Overview of COMET
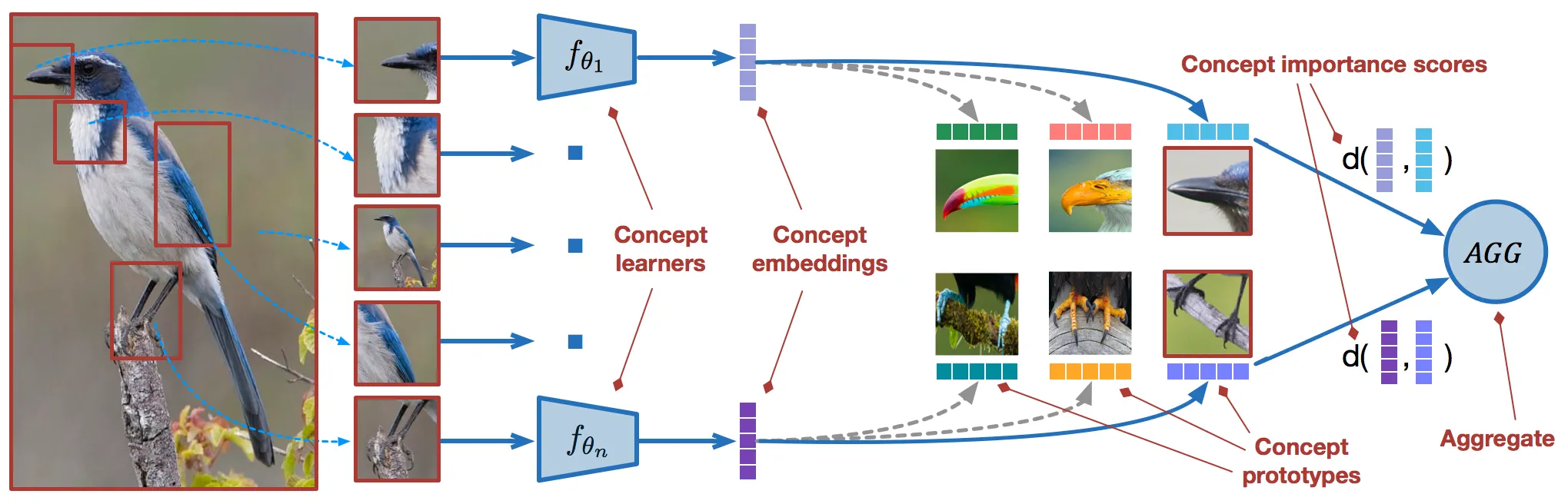
COMET learns a unique metric space for each concept dimension using concept-specific embedding functions, named concept learners, that are parameterized by deep neural networks. Along each high-level dimension, COMET defines concept prototypes that reflect class-level differences in the metric space of the underlying concept. To obtain final predictions, COMET aggregates information from diverse concept learners and concept prototypes. COMET is designed as an inherently interpretable model and assigns concept importance scores to each high-level dimension.
The high-level concepts that are used to guide COMET can be seen as part-based representations of the input and reflect the way humans reason about the world. They can be discovered in a fully unsupervised way, or defined using external knowledge bases. Concepts are allowed to be noisy, incomplete, and/or redundant, and COMET learns which subsets of concepts are important. x
Case studies: image and cell type classification
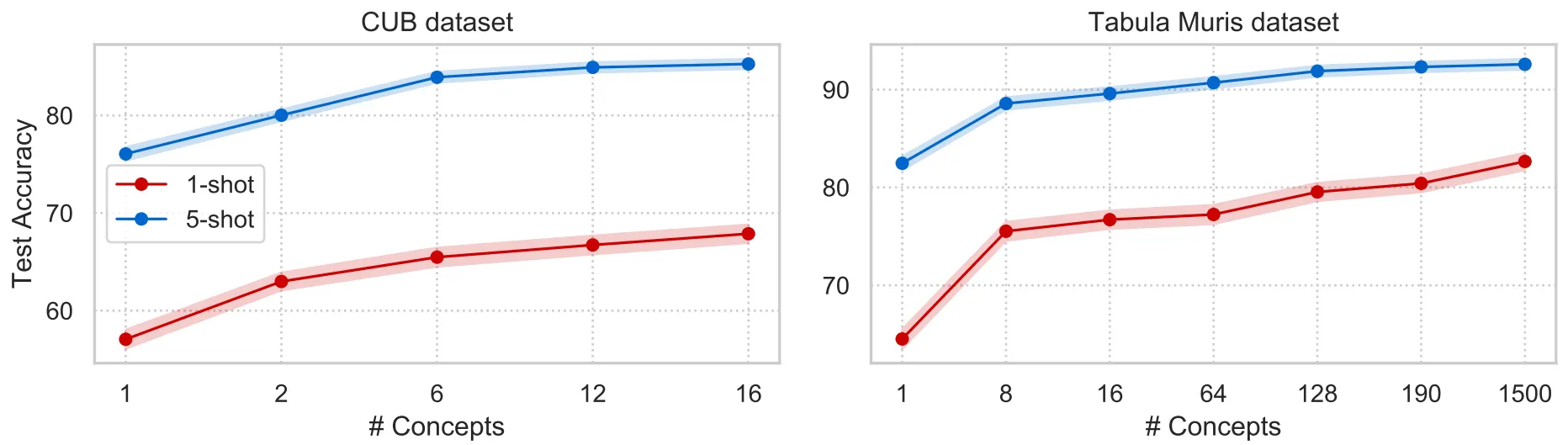
We apply COMET to two diverse tasks: (i) fine grained image classification on CUB-200-2011 dataset of bird species, and (ii) novel cross-organ cell type classification task introduced in our work based on the Tabula Muris dataset. To define concepts on CUB, we use part-based annotations of images (e.g., beak, wing, and tail of a bird). On the Tabula Muris dataset, features correspond to gene expression profiles and we define concepts using Gene Ontology, a resource which characterizes gene functional roles in a hierarchically structured vocabulary. Figure below illustrates the COMET’s performance with the respect to the number of concepts on CUB and Tabula Muris datasets. COMET consistently improves performance when we gradually increase number of concepts even when the concepts are highly redundant and overlapping.
Given a set of query points or an entire class, COMET generates explanations based on the average distance between concept prototype and concept embeddings of all query points of interest. Figure below illustrates an example on the CUB dataset. For each bird species, COMET finds most relevant concepts. For instance, COMET selects ‘beak’ as the most relevant concept for ‘parakeet auklet’ known for its nearly circular beak; ‘belly’ for ‘cape may warbler’ known for its tiger stripes on the belly; while ‘forehead’ is the most relevant feature for ‘belted kingfisher’ known for its shaggy crest on the top of the head.
COMET can also find images that locally resemble the prototypical image and well reflect the underlying concept of interest. Figure below shows an example of images ranked according to the distance of their belly concept embedding to the belly concept prototype.
Novel benchmark dataset
We develop a novel single-cell transcriptomic dataset based on the Tabula Muris dataset that comprises 105,960 cells of 124 cell types collected across 23 organs of the mouse model organism. The features correspond to the gene expression profiles of cells, selected based on the standardized log dispersion. We introduce the evaluation protocol in which different organs are used for training, validation, and test splits, so that a meta-learner needs to learn to generalize to unseen cell types across organs. The dataset along with the cross-organ evaluation splits is available at the link in the table below.
| File | Description |
| tabula-muris-comet.zip | Benchmark Tabula Muris dataset |
Code
A PyTorch implementation of COMET is available on GitHub.
Contributors
The following people contributed to this work: