Generative Modeling Reveals the Connection between Cellular Morphology and Gene Expression
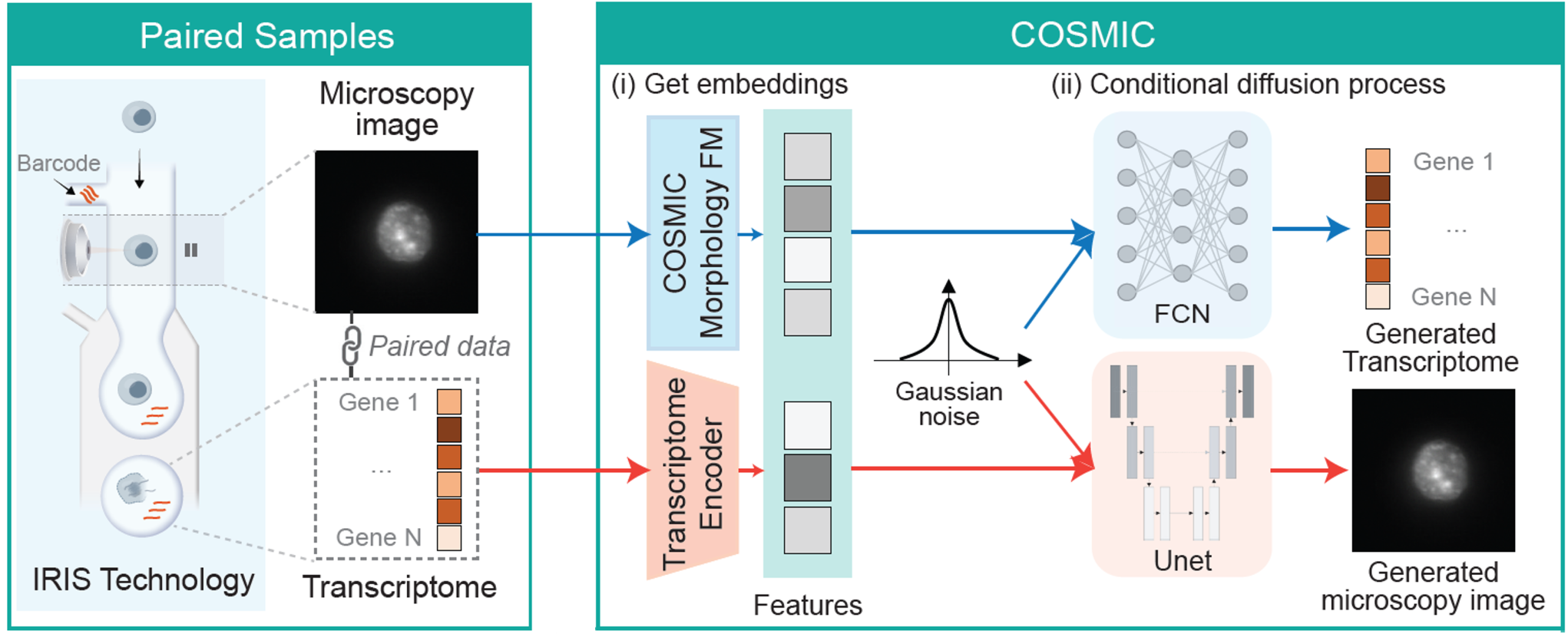
COSMIC (Cross-modal generation between Single-cell RNA-seq and MICroscopy images) is a bidirectional generative framework that quantitatively links single-cell nuclear morphology and gene expression. It captures the bidirectional flow of information between cellular form and gene expression, opening new avenues for mechanistic discovery and predictive modeling in both basic and translational cell biology.
The understanding of how transcriptional programs give rise to cellular morphology, and how morphological features reflect and influence cell identity and function remains limited. This is due in part to the lack of large-scale datasets pairing the two modalities, as well as the absence of computational frameworks capable of modeling their cross-modal structure.
We introduce COSMIC, a bidirectional generative framework that enables quantitative decomposition of transcriptional variance reflected in morphology and morphological variance explained by gene expression. COSMIC accurately modeled cell type identity, as well as continuous dynamics such as cell-cycle progression, establishing a quantitative link between morphological phenotypes and underlying gene expression.
Publication
Generative Modeling Reveals the Connection between Cellular Morphology and Gene Expression
Shuo Wen, Ramon Vinas Torne, Johannes Bues, Camille Lucie Lambert, Nadia Grenningloh, Timothee Ferrari, Elisa Bugani, Joern Pezoldt, Jillian Rose Love, Wouter Karthaus, Bart Deplancke, and Maria Brbić
@article {Wen2026.01.22.700673,
author = {Wen, Shuo and Vi{\~n}as Torn{\'e}, Ramon and Bues, Johannes and Lambert, Camille Lucie and Grenningloh, Nadia and Ferrari, Timoth{\'e}e and Bugani, Elisa and Pezoldt, Joern and Love, Jillian Rose and Karthaus, Wouter and Deplancke, Bart and Brbi{\'c}, Maria},
title = {Generative modeling reveals the connection between cellular morphology and gene expression},
year = {2026},
URL = {https://www.biorxiv.org/content/early/2026/01/24/2026.01.22.700673},
journal = {bioRxiv}
}
Overview of COSMIC
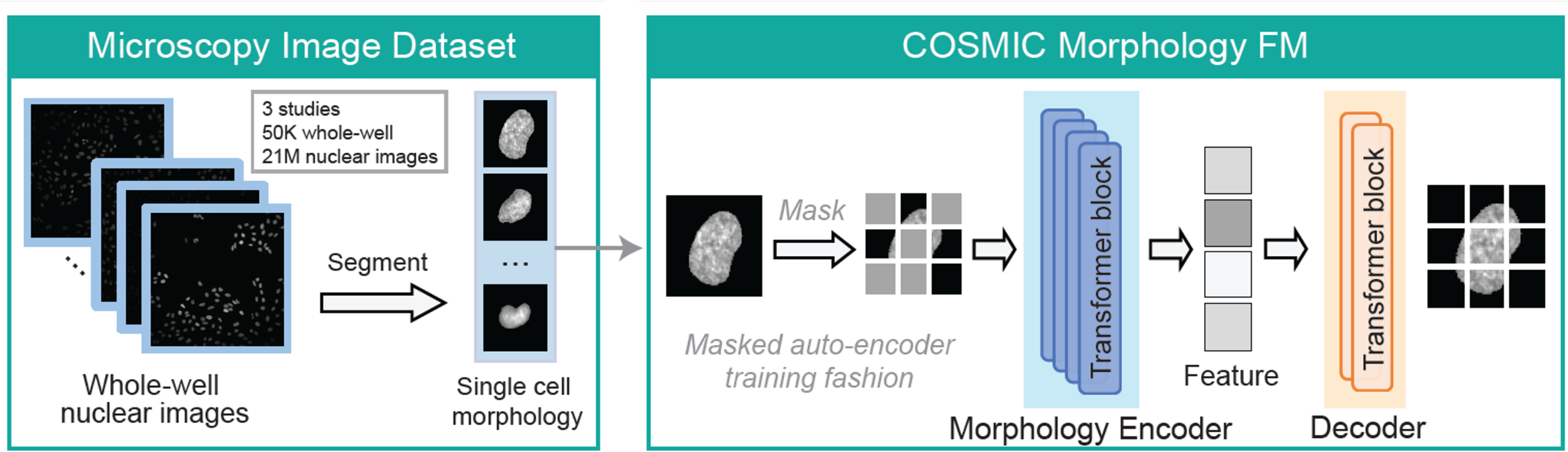
COSMIC captures the relationship between gene expression and cellular morphology by learning to translate from transcriptomes to nuclear images and from nuclear images to transcriptomes. COSMIC builds on a foundation model trained on over 21 million segmented nuclei and couples it with existing transcriptomic embeddings. To enable cross-modal learning, we leveraged a newly generated multimodal dataset acquired using IRIS, a technology that captures high-resolution images and transcriptomes from the same single cells at scale (preprint here).
To build a general-purpose encoder of nuclear morphology, we trained a vision foundation model (FM) on microscopy images of cellular nuclei, referred to as the morphology FM. The model was trained on 21M nuclear images by segmenting and isolating individual nuclear crops from 50K whole-well Hoechst-stained microscopy images from different studies.
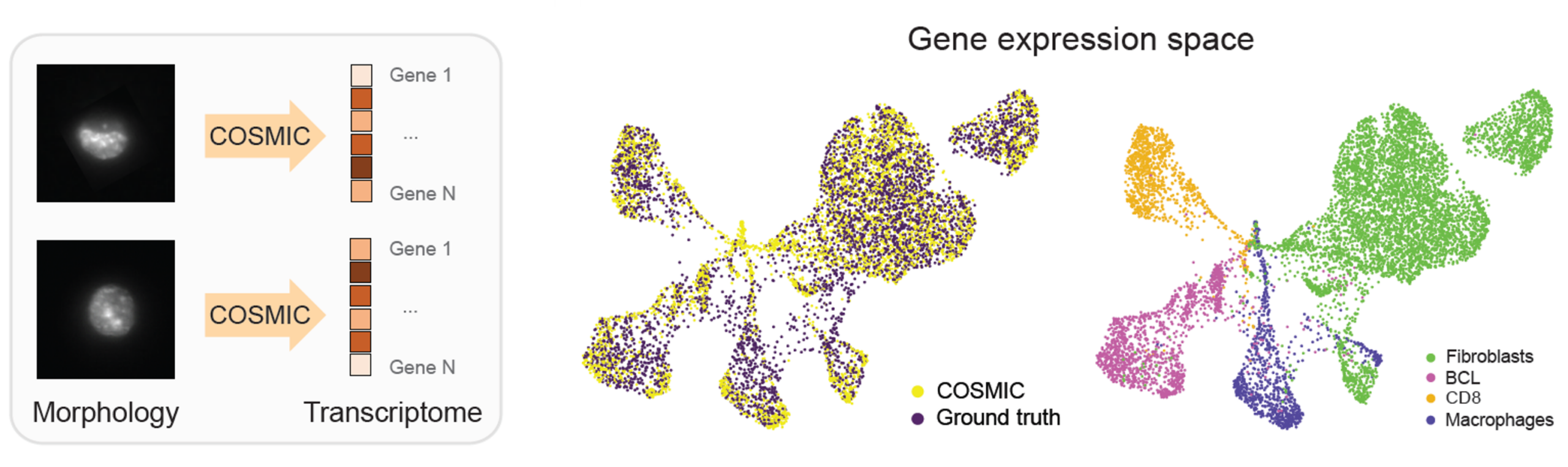
COSMIC generates realistic nuclear images from gene expression
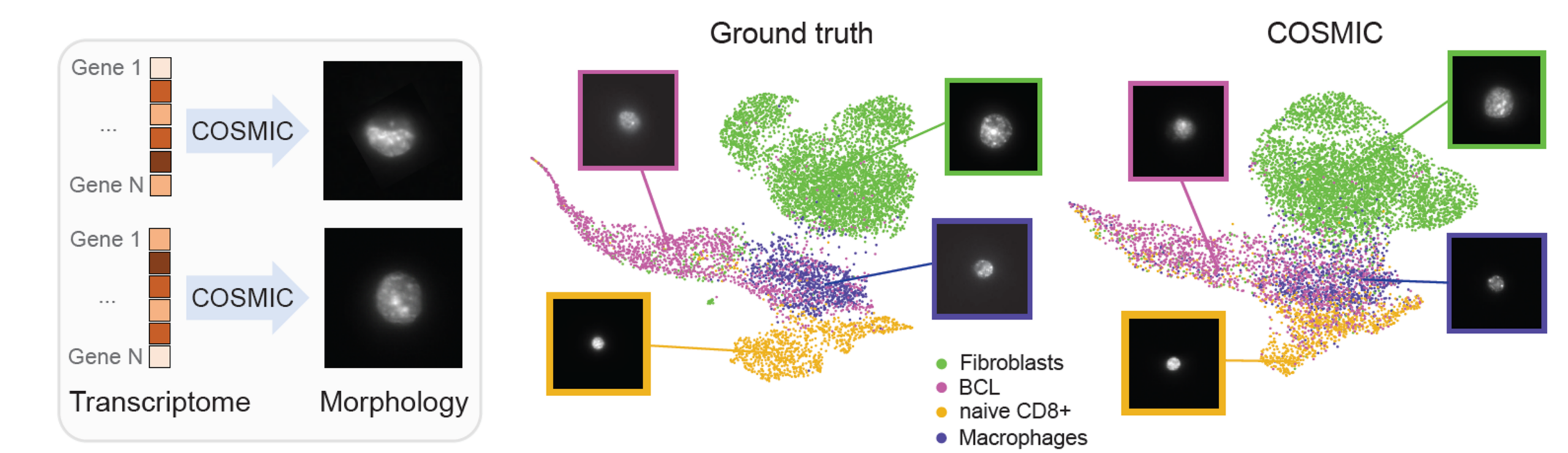
We apply COSMIC on a dataset generated by the IRIS technology that couples high-resolution microscopy with droplet based single-cell RNA sequencing. Specifically, we trained COSMIC on 4,520 paired nuclear image-transcriptome samples of mouse cells, and evaluated it on an independent set of 4,519 samples. For the mouse cells, we profiled embryonic fibroblasts (3T3), macrophage-like cells (RAW), CAR-engineered T cells derived from the A20 B-cell lymphoma model (CAR_A20), and primary naive CD8+ T cells (naive CD8).
COSMIC accurately synthesizes high-resolution nuclear images conditioned on single-cell transcriptomic profiles. Generated images closely match real nuclei in size, shape, and texture, and preserve cell-type-specific morphology. In the embedding space, real and generated nuclei overlap strongly. (BCL stands for the B-cell lymphoma in the following figure.)
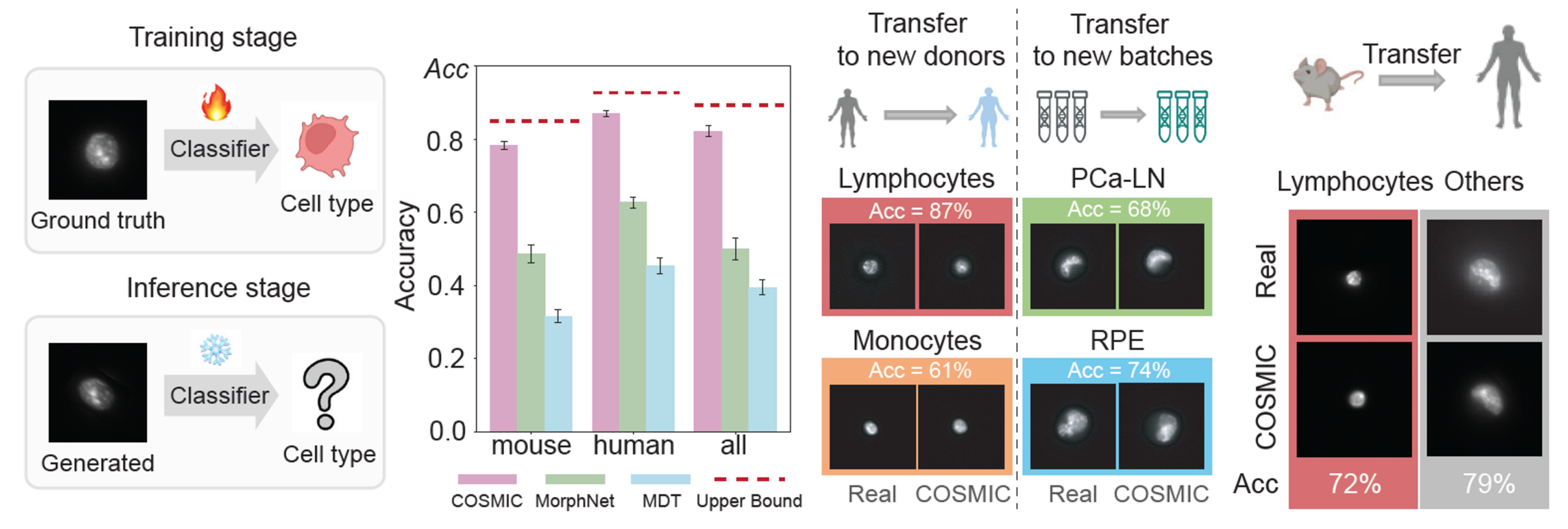
Images generated by COSMIC retain sufficient biological signal to support accurate cell type classification, approaching the performance achieved on real microscopy images. Additionally, COSMIC generalizes to unseen batches, new donors, and even across species.
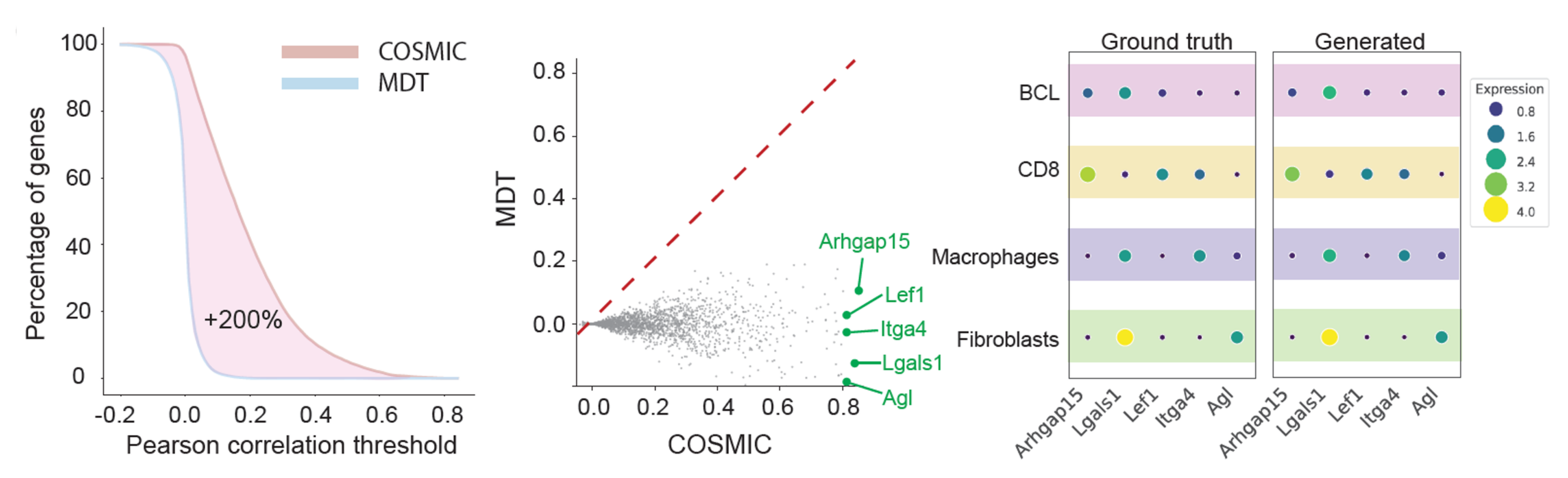
COSMIC predicts transcriptomic profiles from nuclear morphology
We next examine the performance of COSMIC in the reverse direction: predicting transcriptomic profiles from microscopy images of single cells. The dataset and data split are the same as in the previous section.
In this reverse direction, COSMIC infers biologically meaningful transcriptomic profiles directly from nuclear images. Transcriptomes predicted by COSMIC recover the global structure of gene expression space and maintain clear separation between cell types.
At the gene level, COSMIC connects genes to nuclear morphology by identifying a subset of genes whose expression can be robustly predicted from morphology, including cell-type marker genes with high correlation to ground truth. These results demonstrate that nuclear morphology encodes precise information about specific transcriptional programs.
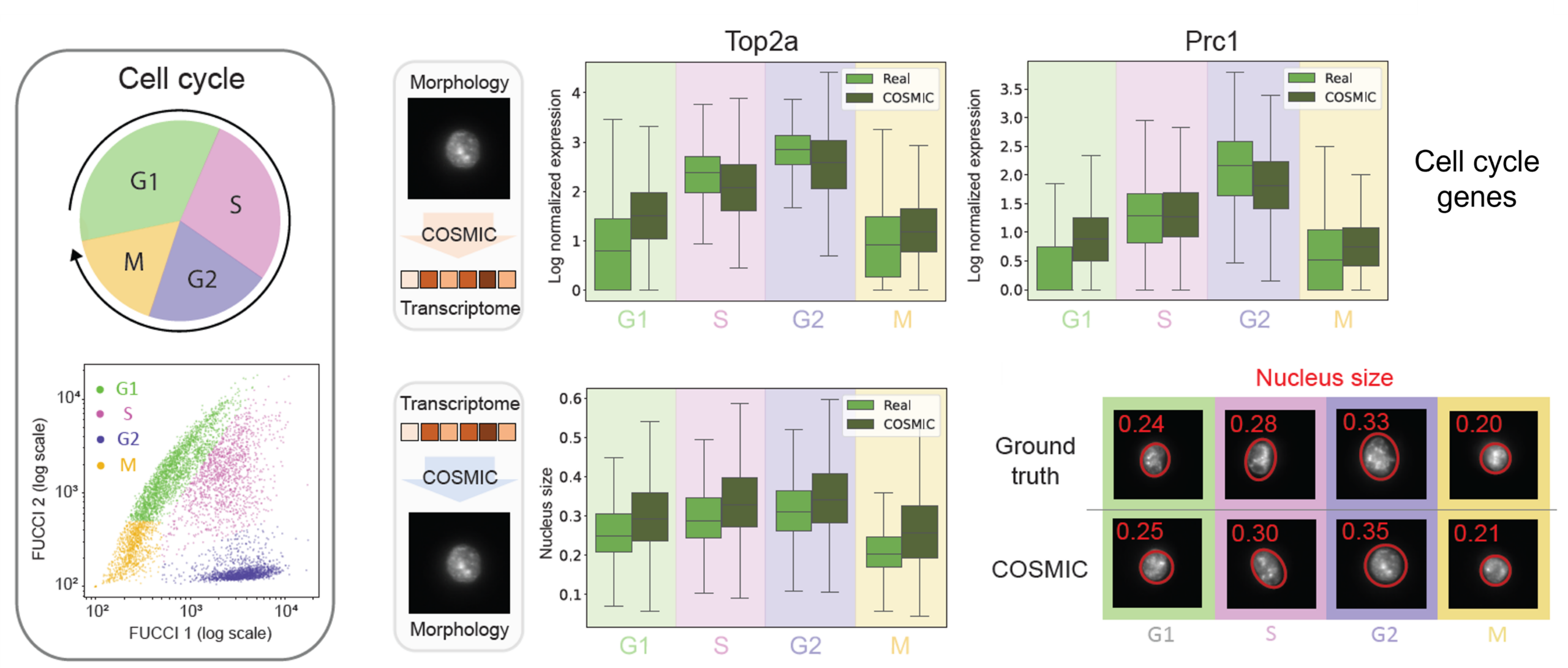
COSMIC captures continuous cell-cycle dynamics
While COSMIC effectively captures cell type identity through relationships between nuclear morphology and transcriptomic profiles, we next explored whether it can also encode biological information beyond discrete cell types and recover continuous biological processes.
Focusing on mouse fibroblasts, we tested whether COSMIC can recover known biological dynamics without supervision, focusing on the cell cycle. The FUCCI reporter signals in this dataset allowed us to explicitly validate the inferred cell cycle profiles. The figure below shows that the predicted transcriptomes reproduce phase-specific expression patterns of canonical cell-cycle genes, while generated nuclear images reflect expected morphological changes across the cell cycle, such as systematic variation in nuclear size.
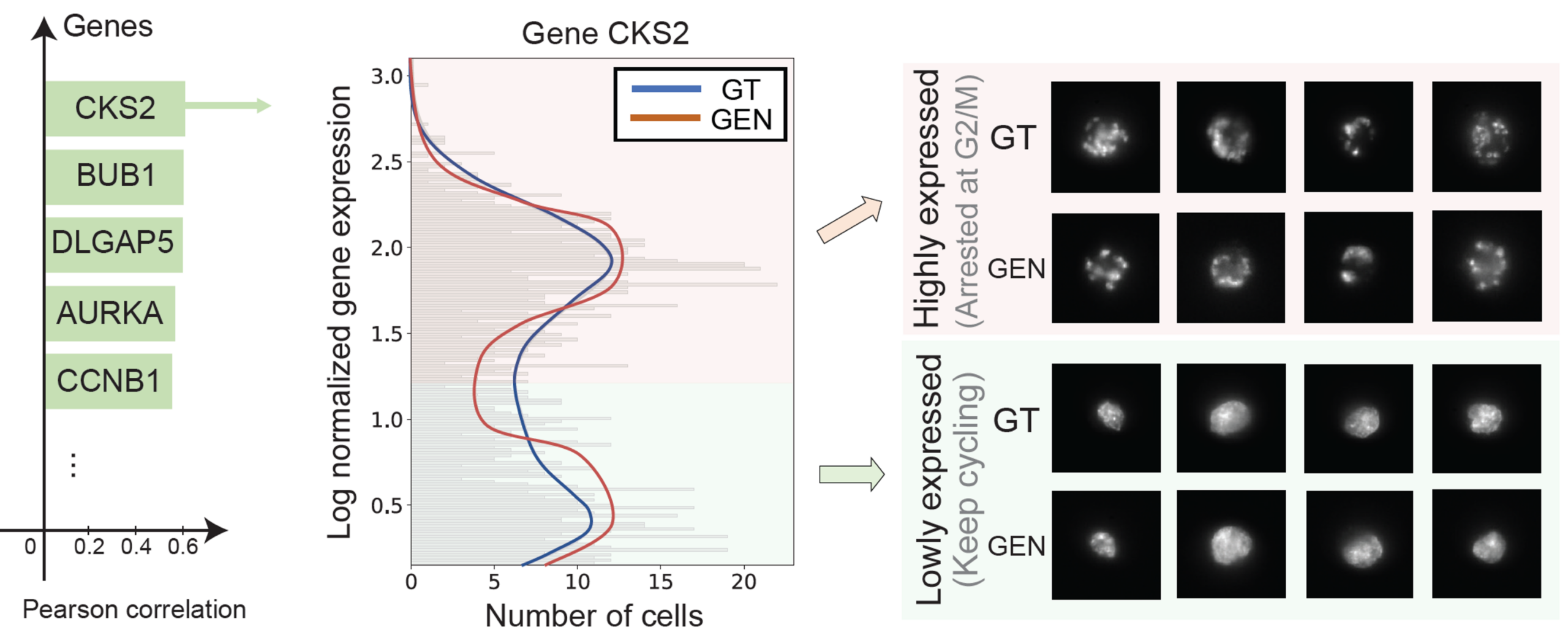
COSMIC identifies morphology-associated genes in cancer
To evaluate whether COSMIC can uncover biologically meaningful morphology–transcriptome relationships in unstructured and clinically relevant contexts, we next applied it to prostate cancer cells. Tumor cells exhibit multiple, overlapping axes of heterogeneity, including proliferation, genomic instability, stress responses, and treatment effects.
COSMIC identified a set of nuclear morphology-associated genes, such as CKS2, BUB1, DLGAP5, AURKA, and CCNB1, which are all well-known regulators of mitosis and drivers of tumor progression. Additionally, COSMIC’s bidirectional structure allowed us to generate synthetic nuclear images conditioned on transcriptomes. These generated images recapitulated the expected morphological differences.
Code
A PyTorch implementation of COSMIC is available on GitHub.
Contributors
The following people contributed to this work:
Johannes Bues
Camille Lucie Lambert
Nadia Grenningloh
Timothee Ferrari
Elisa Bugani
Joern Pezoldt
Jillian Rose Love