HeurekaBench: A Benchmarking Framework for AI Co-scientist
HeurekaBench is a semi-automated framework that turns scientific papers with their code, experimental datasets and findings into open-ended, data-driven research questions for evaluating AI co-scientists. We instantiate our benchmark in the single-cell biology domain and evaluate existing domain-specific LLM-based agents as AI co-scientists.
AI agents can be characterized by a loop consisting of a THINK stage, in which the agent understands the user request and creates a plan; followed by the ACT stage, where it generates an action depending on the plan; and OBSERVE stage, in which it updates the context with the feedback of action execution for the next cycle. Such agents can be used as AI co-scientists by actively generating new hypotheses, designing and evaluating experiments, and iteratively improving through feedback to accelerate scientific discovery. With the rapid progress of AI agents, it is crucial to rigorously design benchmarks to evaluate and further improve their potential as AI co-scientists.
We introduce HeurekaBench, a framework for constructing benchmarks that capture the envisioned role of AI co-scientists: tackling open-ended, data-driven scientific questions through hypothesis-driven exploratory analysis and iterative reasoning.
Publication
HeurekaBench: A Benchmarking Framework for AI Co-scientist
Siba Smarak Panigrahi*, Jovana Videnović*, Maria Brbić.
@article{panigrahi2026heurekabench,
title={HeurekaBench: A Benchmarking Framework for AI Co-scientist},
author={Panigrahi, Siba Smarak and Videnovi{\'c}, Jovana and Brbi{\'c}, Maria},
journal={arXiv preprint arXiv:2601.01678},
year={2026}
}
Co-scientist Task Overview
An AI agent as a co-scientist is expected to handle exploratory research questions grounded in experimental data. The benchmark for evaluating such systems can be formulated as a collection of tasks, where each task consists of a triplet (D, Q, A). Here, D consists of experimental datasets from a specific scientific domain and may include auxiliary files; Q corresponds to an open-ended research question that demands multi-step reasoning over D; and A provides a ground-truth answer to judge agent responses appropriately. This triplet formulation enforces the essential components of real-world co-scientist tasks: (i) authentic experimental datasets, (ii) open-ended research questions requiring workflow-level reasoning, and (iii) scientifically validated answers for evaluation.
HeurekaBench framework
The core idea is to ground benchmark construction in the scientific process itself. We build a multi-LLM pipeline to automatically extract key insights and corresponding multi-step code, which are validated by matching the published results. The validated insights are then used to derive two complementary benchmark formats: (i) open-ended research questions (OEQs), the primary format to measure co-scientist’s performance, as it aligns with exploratory scientific research which requires synthesizing evidence from data, constructing reasoning chains, and articulating conclusions in free-form language and (ii) multiple-choice questions (MCQs), which provide a lightweight and informative proxy for rapid evaluation during agent development.
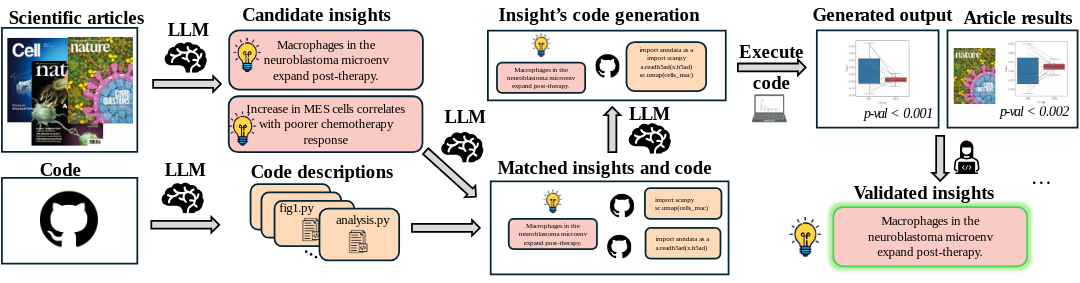
Stage 1: Insights Generation
The first stage of the framework extracts and validates scientific insights from published studies, using their code to retain only reproducible insights. To achieve this, we design a modular pipeline: InsightExtractor proposes candidate insights from the paper, while CodeDescriber converts code scripts into natural language summaries. The outputs of these modules are combined via CodeMatcher, which links the code descriptions and retrieves scripts that could support the insight. Finally, CodeGenerator composes these scripts into a multi-step workflow for each candidate insight.
After generating candidate insight-code workflow pairs, human reviewers run the code to verify each insight’s reproducibility by ensuring that the results, such as figures or statistics, match those reported in the insight and in the study. The output of this stage is a pool of validated insights.
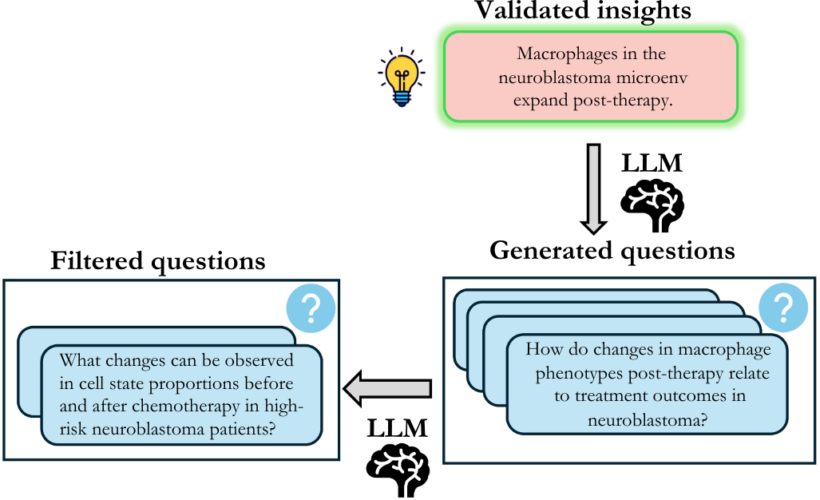
Stage 2: Questions Generation
For each validated insight, we generate two question types, as described before: (i) OEQs and (ii) MCQs. The generation process for both follows a similar pipeline, differing only in the prompting strategies for automatic generation and the subsequent filtering steps. We employ few-shot prompting to generate (Q, A) pairs for each insight. For MCQs, we emphasize creating challenging distractors that capture plausible misinterpretations or common analytical errors, ensuring that correct answers require a genuine understanding of the data. OEQs, on the other hand, are intentionally less specific, allowing multiple approaches to reach the correct answer. Following the generation, questions undergo a two-stage filtering process: (i) automatic filtering to remove easy questions solvable using LLMs’ pretraining knowledge and (ii) manual review to remove hallucinations, duplicates, and questions based on non-validated parts of the insights.
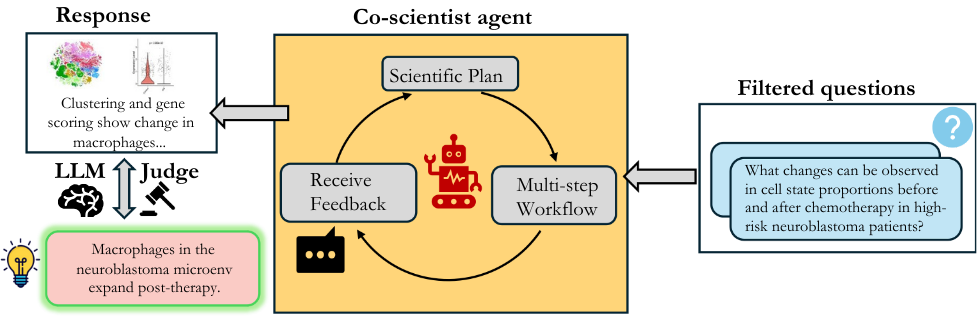
Stage 3: Scientifically-grounded Evaluation
Evaluation of OEQs poses interesting challenges. Agent analyses on these questions may uncover additional conclusions beyond the annotated ground truth. Moreover, in natural-language responses, an agent could rely on prior knowledge from the literature rather than exploring the dataset. Therefore, the evaluation must go beyond surface-level matching. We propose a new evaluation scheme that instructs the LLM-Judge evaluator to first decompose both the agent’s and the ground-truth’s answers into atomic facts (e.g., conditions, trends, conclusions) and then assess overlap across complete, partial, and missing facts. The evaluator assigns a correctness score between 1 and 5. An agent receives the highest score only if all ground-truth facts are present and no contradictions occur, while additional non-conflicting findings are not penalized (the complete evaluation rubric is available in Appendix B of the paper). This evaluation design rewards dataset-backed findings rather than factual recall and thus aligns with the use of AI co-scientists.
For MCQs, we report accuracy as the primary metric. Although all choices are generated solely from validated insights, some options marked as incorrect may still appear scientifically plausible due to being LLM-generated. To account for such scenarios, we also use precision and recall.
scHeurekaBench: Benchmark for AI Co-scientist in Single-cell Biology
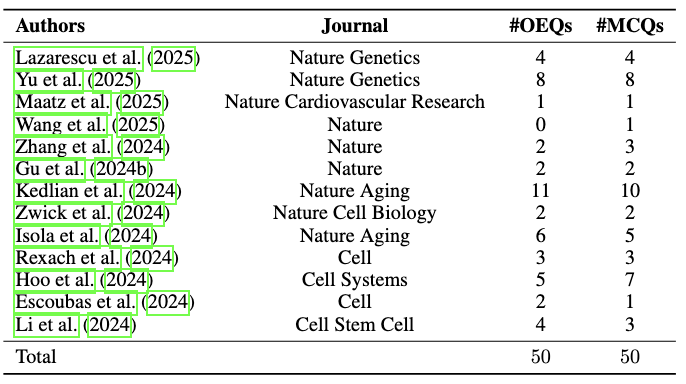
We instantiate the HeurekaBench framework in single-cell biology, particularly for studies on the analysis of scRNA-sequencing datasets. First, we curate a pool of 22 papers published in Nature and Cell journals in 2024 and 2025. We then applied our insight-generation pipeline to produce 10 candidate insights per paper, retaining only those that could be validated. This process yielded a final pool of 41 validated insights across 13 papers. Finally, using our question-generation pipeline, we derived 50 OEQs and 50 MCQs from the validated insights to construct sc-HeurekaBench. We also create sc-HeurekaBench-TU, a set of 12 OEQs that explicitly rely on specific tool usage (e.g., scenic, cellchat, cellphoneDB), to evaluate the importance of a retriever component within agents.
Overall Results
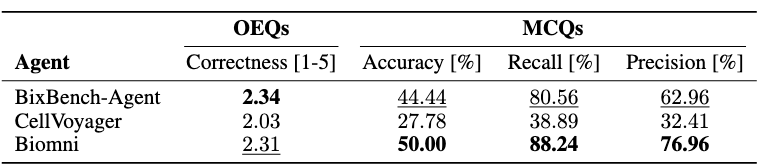
We benchmark three state-of-the-art agents for single-cell biology using our sc-HeurekaBench: Biomni [1], CellVoyager [2], and BixBench-Agent [3], all built with Claude-4-Sonnet LLM. Since BixBench-Agent crashed on large datasets, we report below the results for questions on datasets smaller than 750 MB, which contain 22 OEQs and 18 MCQs on which all agents could run. We term this benchmark as sc-HeurekaBench-Lite.
We observe that BixBench-Agent and Biomni outperform CellVoyager in both formats, indicating that a more flexible agent loop can build robust workflows. Biomni also includes more domain-specific tools and databases than other agents. We inspected the CellVoyager outputs, which revealed limited code-fixing capabilities and difficulty incorporating multiple feedback at each step. Further, the number of steps needs to be prespecified, which limits the workflow from finishing and producing an appropriate answer. In subsequent ablations, we focus on Biomni as the only agent that can run the entire benchmark at high performance.
Ablations on Planner component within agents
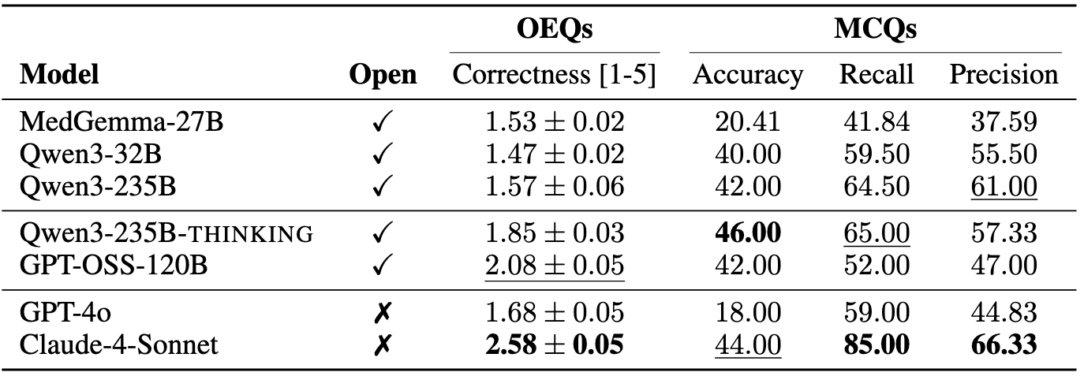
The planner component within the agent is responsible for generating the initial plan and subsequently receiving feedback from the environment (e.g., results, error messages) to modify it. In Biomni, the planner can decide to generate either a plan or code actions as required. We compare a range of open- and closed-source LLMs across model sizes, families, and reasoning style.
We observe that closed-source Claude-4-Sonnet achieves the highest overall performance across OEQs and MCQs. Particularly in OEQs, it outperforms the second-best model by a significant margin, highlighting the benefits of closed-source frontier models. Within the Qwen-model family, performance consistently improves with increasing model size, and the thinking variant provides additional gains. Among open-source models, the best-performing model was GPT-OSS-120B, specifically designed for use within agentic workflows.
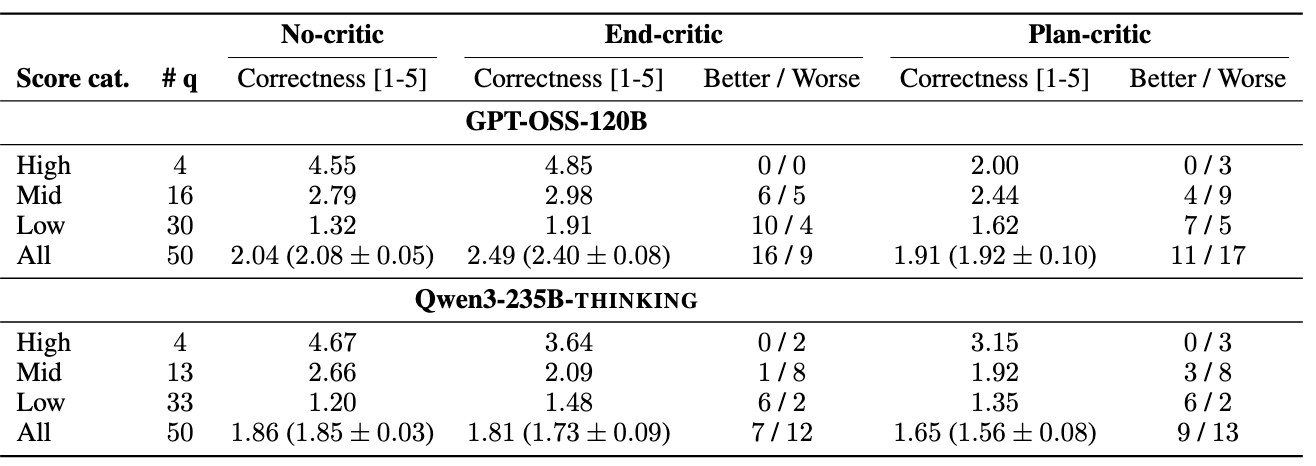
Ablations on Critic component within agents
The critic LLM provides critical recommendations on the outputs at different stages of the agent loop. In our experiments, we compare different positions for the critic module. No-critic denotes the absence of a critic LLM. Additionally, we compare Plan-critic and End-critic, where the critic is present either immediately after the planner drafts an initial plan or at the end of analysis, when the planner decides to exit. We categorize the original scores into high-, mid-, and low-performing questions. Alongside, we also count the number of questions that achieved better or worse scores with different critics.
The extent of benefit depends on the underlying LLM. Furthermore, the content and quality of feedback are influenced by the critic LLM and, more importantly, the responses generated by the planner, which adds stochasticity to the final answers and workflows generated by agentic systems with a critic. In particular, for GPT-OSS-120B, the End-critic yields consistent gains in performance, raising average correctness scores up to 2.49 (close to 2.58 with Claude-4-Sonnet), with the strongest effect on low-scoring questions.
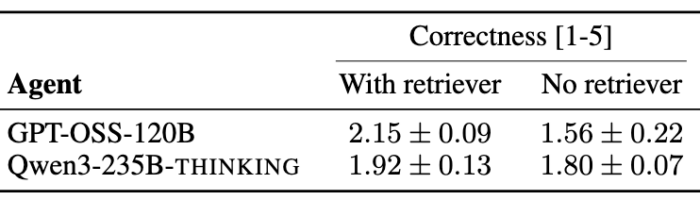
Ablations on Retriever component within agents
The retriever module is an LLM tasked with selecting the appropriate tools, software, and databases from the environment relevant to solving the task before the initial plan is created. This intermediate step avoids overwhelming the planner with the extensive set of tools available. We evaluate agents and sc-HeurekaBench-TU, where the agent requires access to domain-specific tools to provide a well-formed answer.
We observe a significant drop in correctness scores across multiple agent runs, indicating that without the retriever, the agent is unable to choose the proper set of tools and submits a suboptimal response.
Analysis: Failure modes in Open-source LLM-based AI Co-scientist
One of the essential reasons for designing sc-HeurekaBench is to understand the failure modes of current open-source LLM-based agents used as AI co-scientists. We manually analyze agent responses to provide preliminary insights below:
- Incorrect scientific skills: where the agent recalls scientific knowledge from pre-training, e.g., instead of calling a tool for gene set enrichment (pathway) analysis, it recalls a random number of canonical gene markers for specific pathways (or from the MCQ options) and identifies their mean expression to suggest if a pathway is activated.
- Lack of dataset exploration: in several cases, the agent does not explore all metadata columns and thus cannot find suitable information, even though it could be identified in the given datasets.
- Insufficient environment exploration: although the agent uses only a few of the retrieved tools, it doesn’t explore or consider additional components of the environment that could enable a more informed, holistic response.
- Hallucinations: the agent directly generates an answer without any or incomplete analyses, resulting in a response that lacks meaningful data-driven conclusions.
- Other: includes the agent (i) writing large code blocks instead of small, step-wise code snippets, (ii) being unable to take code execution errors into account, and (iii) relying on known literature to eliminate options from MCQs (which actively goes against the idea of data-driven discovery)
Equipping open-source LLMs with these skills through appropriate post-training approaches can significantly enhance their adoption as agents for scientific discovery.
References
[1] Kexin Huang et al. Biomni: A general-purpose biomedical AI agent. bioRxiv, 2025.
[2] Samuel Alber et al. Cellvoyager: AI compbio agent generates new insights by autonomously analyzing biological data. bioRxiv, 2025.
[3] Ludovico Mitchener et al. Bixbench: a comprehensive benchmark for llm-based agents in computational biology. arXiv preprint arXiv:2503.00096, 2025.
Code
Code to evaluate AI co-scientists using scHeurekaBench and to reuse the HeurekaBench framework to create benchmarks for other scientific domains is available on GitHub.
Contributors
The following people contributed to this work: