Tissue Reassembly with Generative AI
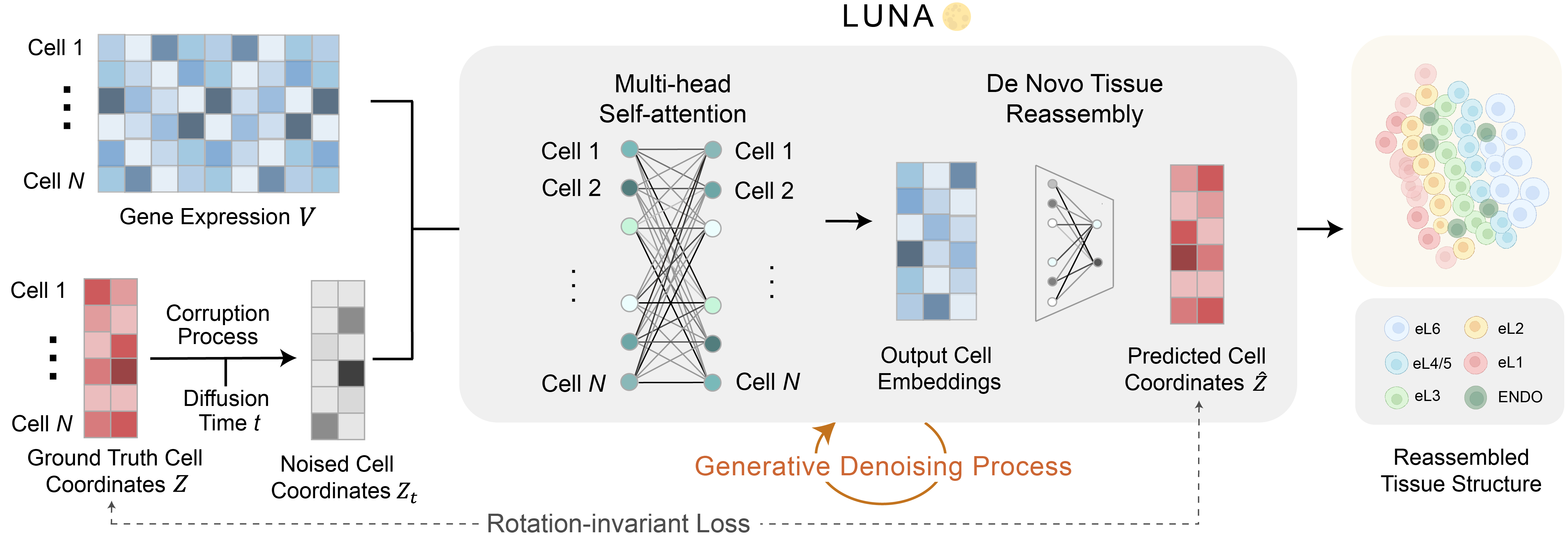
LUNA is a generative AI model that reconstructs tissue structures from gene expressions of cells by learning spatial priors over existing spatially resolved datasets. It captures the complex spatial interrelationships of cells within tissues, enabling de novo reconstruction of cell locations.
Single-cell RNA sequencing (scRNA-seq) technologies have enabled high-throughput profiling of cellular transcriptomes, revealing unique transcriptional profiles within individual cells and advancing our understanding of cellular diversity. Despite this progress, a key limitation of scRNA-seq is the loss of spatial context, which is important for understanding how cells interact with their environment. Spatially resolved sequencing technologies aim to address this issue, but they are constrained by the number of genes that can be measured.
We develop LUNA (Location reconstrUction using geNerative Ai), a generative AI model that reassembles complex tissue structures from gene expressions of cells by learning spatial priors over spatial transcriptomics datasets. LUNA learns cell representations that capture cellular interactions globally and locally across the entire tissue slice, enabled via an attention mechanism that takes into consideration interactions across all cells. LUNA operates as a diffusion model – during training it learns to denoise corrupted cell coordinates, while during inference it starts from random noise and reconstructs physical locations of cells de novo solely from their gene expressions.
Paper
Tissue reassembly with generative AI.
Tingyang Yu, Chanakya Ekbote, Nikita Morozov, Jiashuo Fan, Pascal Frossard, Stéphane d’Ascoli, Maria Brbić.
bioRxiv, 2025.
Overview of LUNA
During training, LUNA takes as input a gene expression matrix along with the corresponding cell coordinates from spatial transcriptomics data. During the inference phase, LUNA takes as input gene expressions without any spatial information and predicts cell locations .
LUNA consists of a multi-head self-attention mechanism that allows each cell to focus on gene expressions of specific cells when making predictions. The cell embeddings are then mapped from the latent space to the physical space via a fully connected layer. LUNA is highly scalable, with linear time and memory complexity relative to the number of cells.
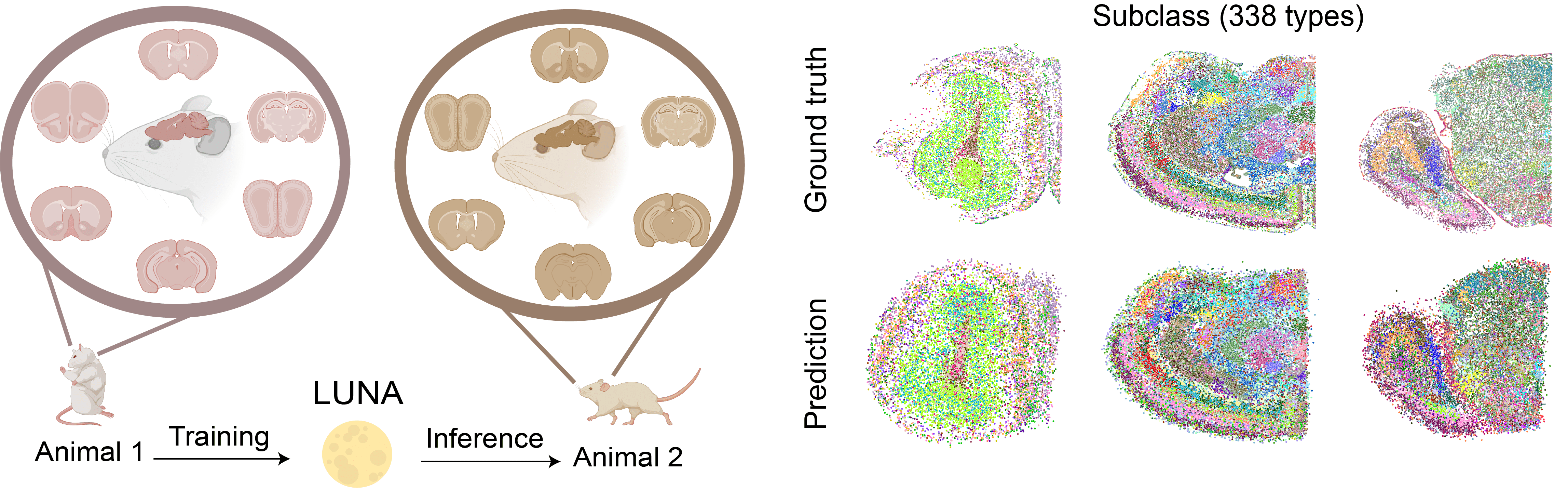
LUNA reconstructs the whole mouse brain atlas
We applied LUNA on a whole mouse brain of the Allen Brain Cell (ABC) MERFISH mouse brain atlas. We trained LUNA on 2.85 million cells across 147 slices from one mouse. We then applied it to generate locations for every cells of the whole mouse brain of another mouse, never seen during model training, consisting of 1.23 million cells and 338 identified subclasses across 66 slices.
LUNA accurately generated the complex architecture of the whole mouse brain conditioned only on the gene expressions of cells. The predictions generated by LUNA closely align with the ground truth cell locations across 11 major regions identified in the ABC atlas, despite their distinct structural characteristics. For example, LUNA accurately captured the circular structure of the olfactory bulb region, the layered organization of the isocortex region, and the anatomical separation between the brain stem and cerebrum.
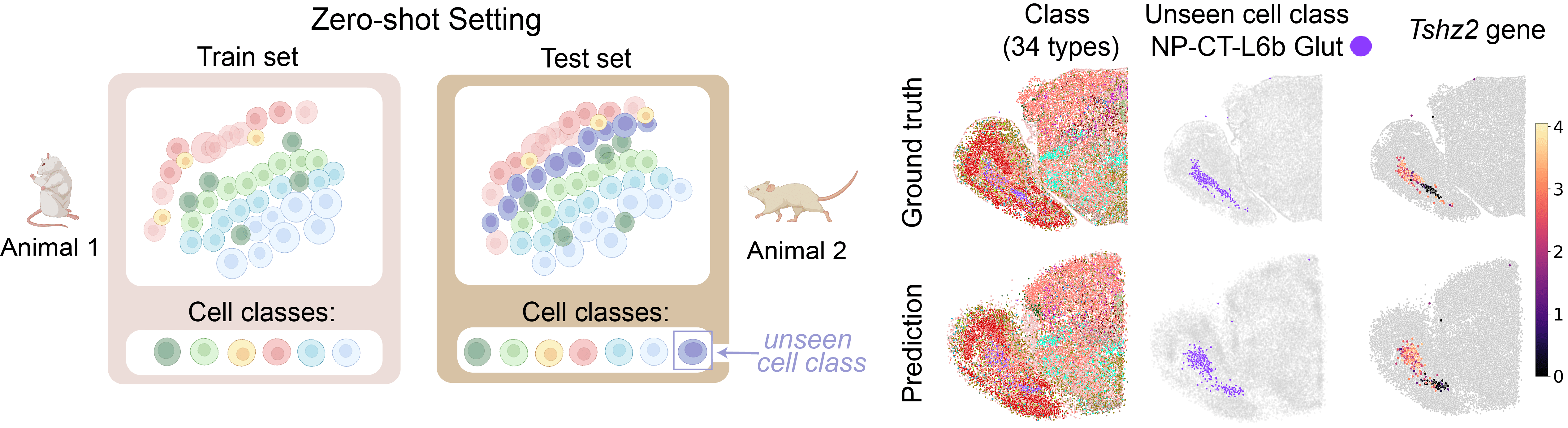
LUNA generalizes to unseen cell classes
We next evaluated zero-shot capability of the model to predict the spatial locations of cells from classes unseen during model training without any additional training or fine-tuning of the model. We trained LUNA on all slices from the Animal 1 in the ABC atlas, excluding NP-CT-L6b Glut cells (n=69,641). The NP-CT-L6b Glut class was randomly chosen from 34 cell classes identified in the dataset.
We then applied LUNA to predict tissue structures for all cell classes in Animal 2, including unseen NP-CT-L6b Glut cells (n=29,262) that have never been seen during model training. The predictions closely matched the ground truth locations in all cell classes. Across different slices, LUNA correctly positioned NP-CT-L6b Glut cells within the spatial architecture of the tissue. Within the unseen NP-CT-L6b Glut class, LUNA also successfully positioned cells according to the expression patterns.
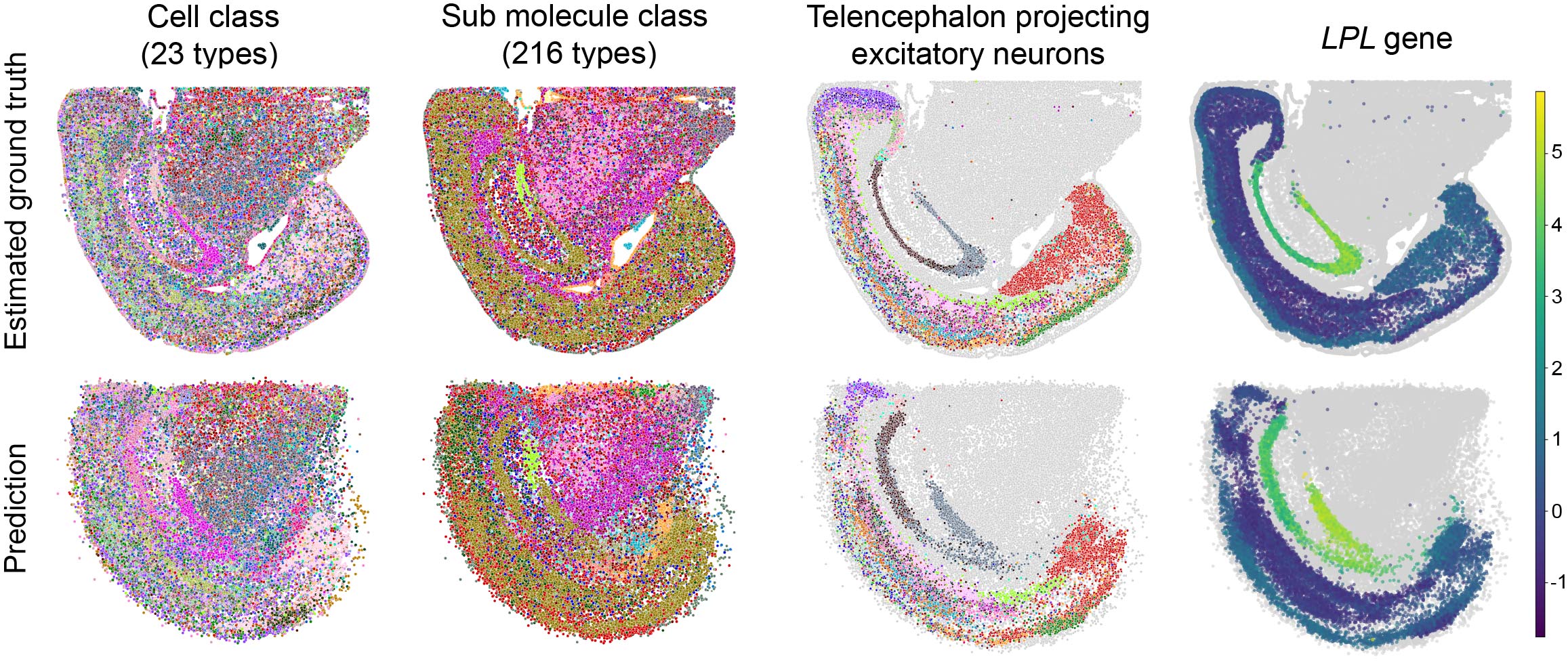
LUNA reassembles scRNA-seq CNS atlas de novo
We next applied LUNA for de novo generation of tissue structures of 1.08 million dissociated single cells across 13 coronal slices from the mouse central nervous system (CNS) scRNA-seq atlas. We used the model trained on the ABC MERFISH mouse brain atlas to predict spatial locations for cells in the scRNA-seq mouse CNS atlas. Since the scRNA-seq atlas lacks spatial information, we validated LUNA’s performance by using estimated cell locations derived from the integration of the scRNA-seq atlas with the STARmap PLUS CNS spatial atlas.
LUNA’s predictions aligned closely with cell locations estimated through the integration data with the STARmap PLUS atlas at both the coarse cell class level and the finer submolecular class level. Additionally, we examined LUNA’s predictions for specific neuronal subtypes including telencephalon projecting excitatory neurons, along with their respective sub-molecular classes. The results indicate that LUNA not only accurately placed cells across major cell classes but also precisely predicted the spatial relationships of complex sub-molecular classes within these neuronal cells.
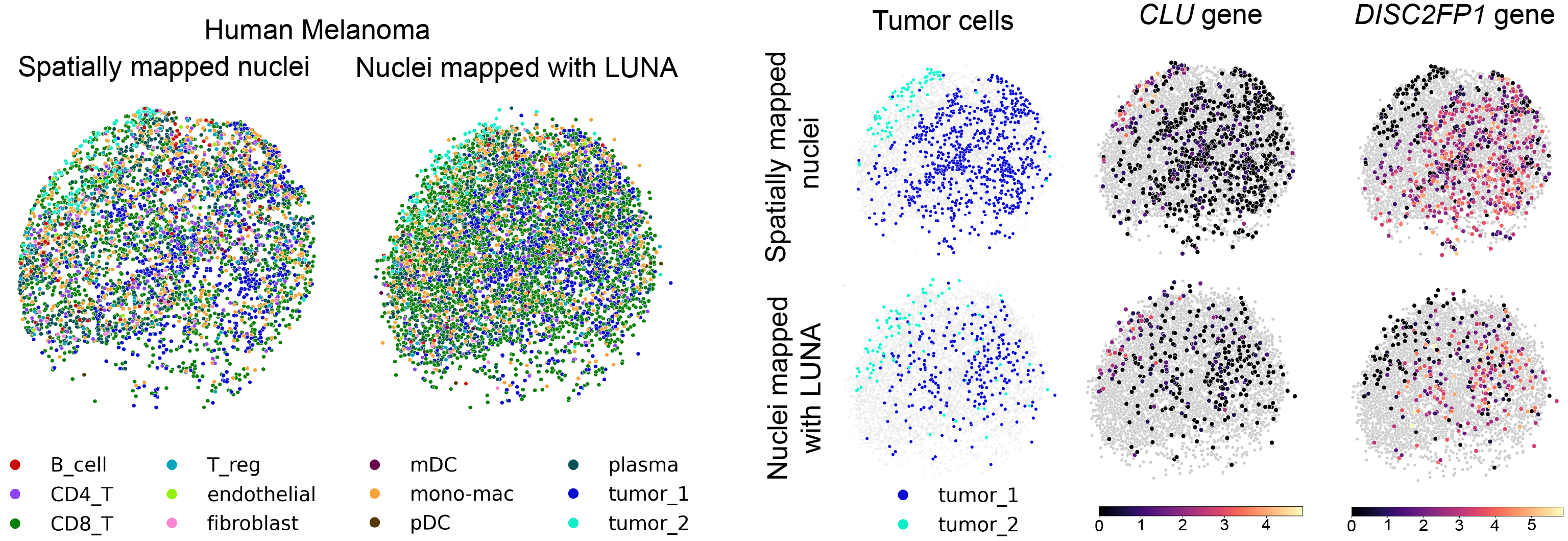
LUNA infers locations of spatially unmapped nuclei in the Slide-tags data
LUNA can be used to predict the tissue locations of nuclei lost during cell profiling with Slide-tags technology. Slide-tags enables profiling single-cell and spatially resolved transcriptome by tagging nuclei with spatial barcode oligonucleotides derived from DNA-barcoded beads with known positions and then using tagged nuclei as an input to single-nucleus profiling assays. However, many nuclei are lost during the barcoding process due to a combination of dissociation and microfluidic losses. To compensate for the sparsity in Slide-tags data, we applied LUNA to assign spatial locations to the spatially unmapped nuclei in the Slide-tags data.
We applied LUNA to a human metastatic melanoma sample obtained with Slide-tags technology. LUNA successfully enriched the dataset from 4,804 cells to the total of 6,466 spatially mapped cells. Notably, LUNA correctly placed two tumor subpopulations into spatially distinct compartments according to predicted cell class annotations.
Datasets
Allen Brain Cell (ABC) MERFISH mouse brain atlas is from Zhang et al. Nature ’23
scRNA-seq Mouse Central Nervous System Atlas is available at the Single Cell Portal
Slide-tags Dataset for the human melanoma tissue is available at the Broad Institute Single Cell Portal under the following accession numbers: SCP2171
Code
A PyTorch implementation of LUNA is available on GitHub.
Contributors
The following people contributed to this work: