Systema: A Framework for Evaluating Genetic Perturbation Response Prediction Beyond Systematic Variation
We evaluate the capabilities of existing perturbation response prediction methods and find that their high predictive scores are largely driven by systematic biases. We introduce Systema, an evaluation framework that emphasizes perturbation-specific effects and identifies predictions that correctly reconstruct the perturbation landscape.
Understanding the effects of perturbations on single cells is essential for advancing functional genomics and therapeutic discovery. To navigate the complex combinatorial landscape of genetic perturbations, computational approaches have been developed to predict transcriptional outcomes of genetic perturbations that were never experimentally tested. However, despite the strong performance reported for these methods, their ability to infer the effects of unseen perturbations remains an open question. Here, we benchmark commonly used single-cell perturbation datasets and observe that simple baselines perform comparably to existing methods. We show that this result can be largely explained by systematic variation, i.e., systematic differences between perturbed and control cells caused by selection biases in the perturbation panel or underlying biological factors. To gain insight into the biology captured by perturbation response prediction methods, we introduce Systema, a new evaluation framework that i) mitigates systematic biases by focusing on perturbation-specific effects, and ii) provides an interpretable readout of the methods’ ability to reconstruct the perturbation landscape. Systema reveals that while generalizing to unseen perturbations remains a substantial challenge, some methods can partially recover the effects of perturbations targeting functionally coherent gene groups. Systema can help differentiate predictions that merely replicate systematic effects from those that capture biologically informative perturbation responses.
Publication
Systema: A Framework for Evaluating Genetic Perturbation Response Prediction Beyond Systematic Variation
Ramon Viñas Torné, Maciej Wiatrak, Zoe Piran, Shuyang Fan, Liangze Jiang, Sarah A. Teichmann, Mor Nitzan, Maria Brbić.
@article{vinas2025systema,
title={Systema: A Framework for Evaluating Genetic Perturbation Response Prediction Beyond Systematic Variation},
author={Vinas Torne, Ramon and Wiatrak, Maciej and Piran, Zoe and Fan, Shuyang and Jiang, Liangze and A. Teichmann, Sarah and Nitzan, Mor and Brbic, Maria},
journal={Nature Biotechnology},
year={2025},
}
Simple baselines perform comparably to existing perturbation response prediction approaches
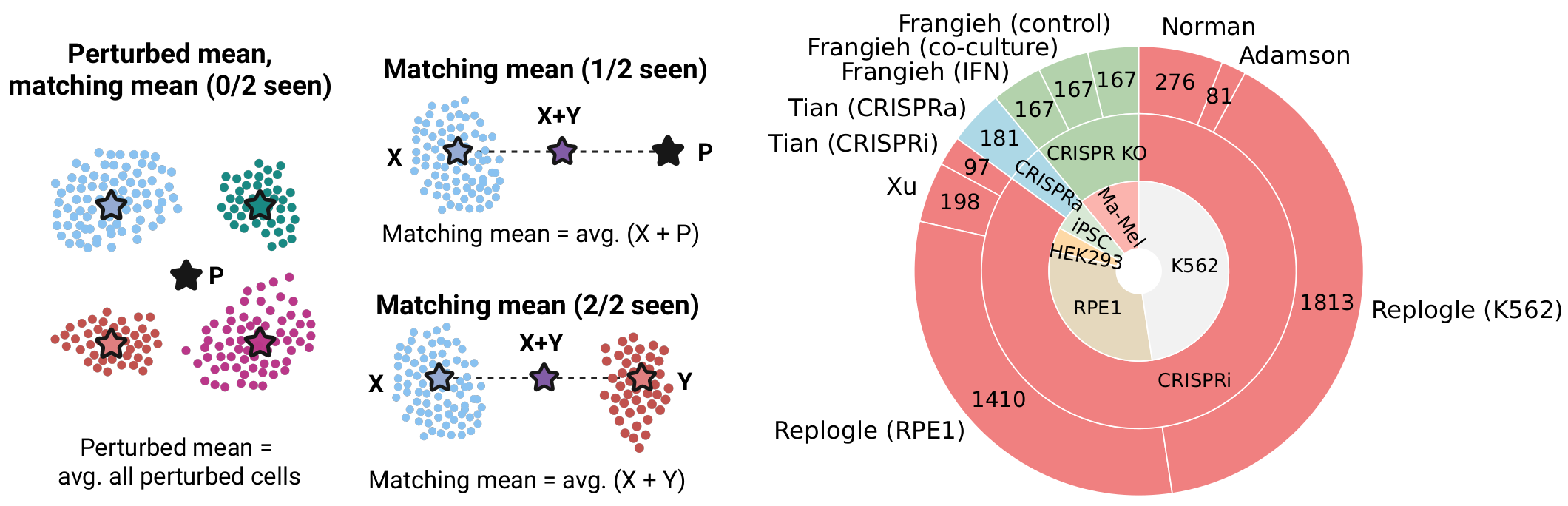
We benchmarked established perturbation response methods on data from ten single-cell perturbation datasets. We designed two simple non-parametric baselines that capture average perturbation effects: i) average expression across all perturbed cells, that we refer to as perturbed mean, and ii) average expression across matched post-perturbation profiles for combinatorial perturbations, referred to as matching mean. Specifically, the matching mean for perturbation X+Y is the average of the X and Y centroids, and if X or Y are unseen at train time, their centroid is replaced by the perturbed mean.
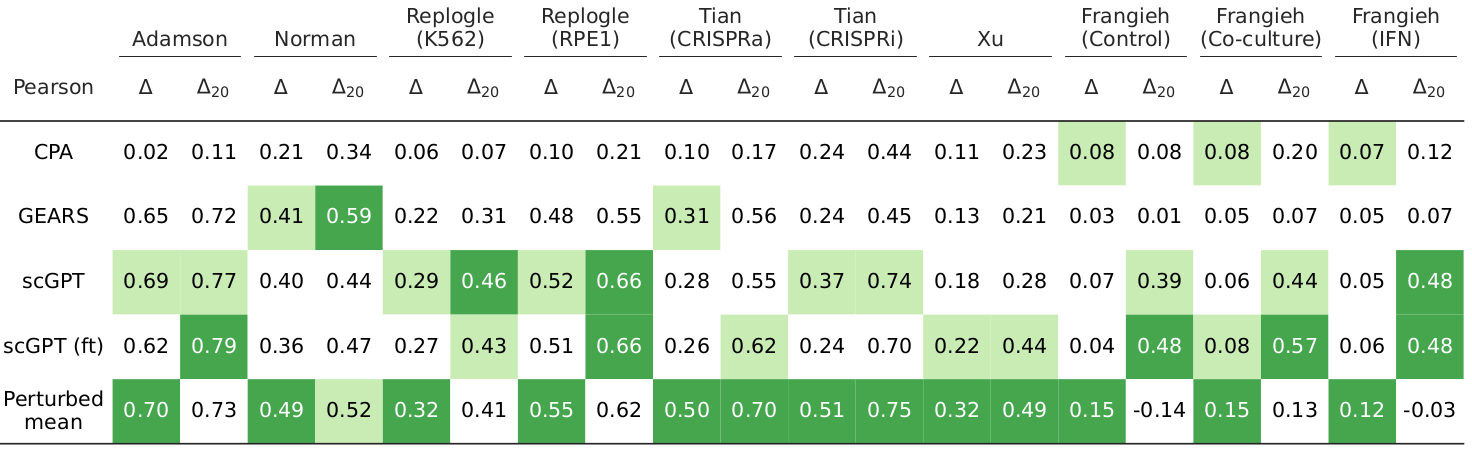
We studied to what extent methods can predict the transcriptional changes induced by unseen genetic perturbations, defined as the average difference in gene expression between perturbed cells subjected to a given perturbation and control cells (i.e., the average treatment effect). We included two simple baselines (e.g. average expression of perturbed cells; perturbed mean) and found that they performed comparatively or outperformed state-of-the-art methods across different datasets and evaluation metrics.
Systematic differences between control and perturbed cells lead to high predictive scores
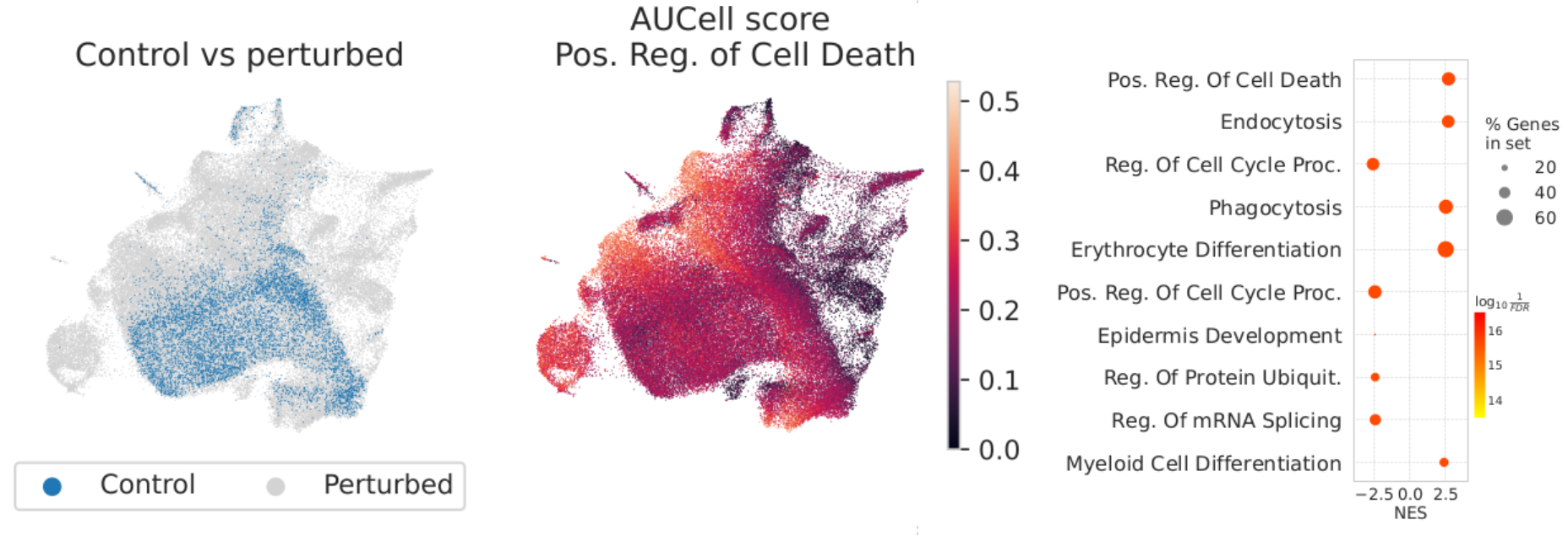
We hypothesized that the comparatively high predictive scores of these baselines reflect systematic differences between perturbed and control cells. The presence of systematic differences that explain variation in the gene expression of single cells is prevalent across datasets and can greatly affect downstream analyses. For example, systematic variation can lead to overestimated predictive performance of perturbation response models when they primarily capture the average perturbation effect, obscuring their ability to generalize to novel perturbations. In perturbation datasets, systematic differences between perturbed and control cells may be explained by potential selection biases, confounding variables, or underlying biological factors. Systematic variation may still arise when responses that are biological in origin but systematic in effect, such as stress response or cell cycle arrest, occur broadly across many perturbations.
Standard reference-based metrics are susceptible to systematic variation
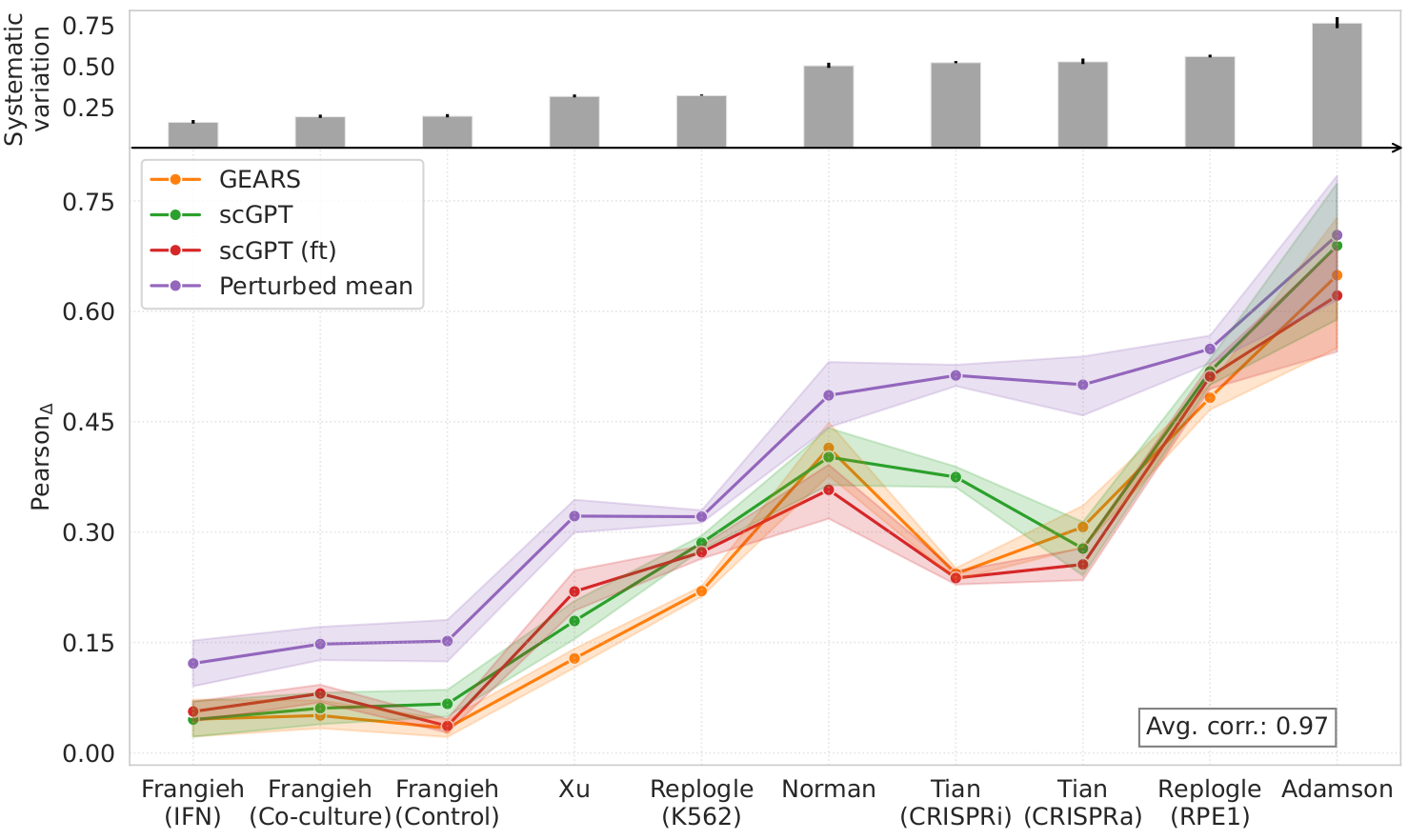
We next studied to what extent standard evaluation metrics are affected by systematic differences between perturbed and control cells. In presence of systematic variation, cells subjected to different perturbations may consistently exhibit similar gene expression shifts with respect to the population of control cells. To quantify systematic variation, we computed the distribution of cosine similarities between perturbation-specific shifts and the average perturbation effect. High cosine similarity indicates that the transcriptional responses to different perturbations are aligned in a similar direction, suggesting shared, possibly non-specific, shifts in gene expression.
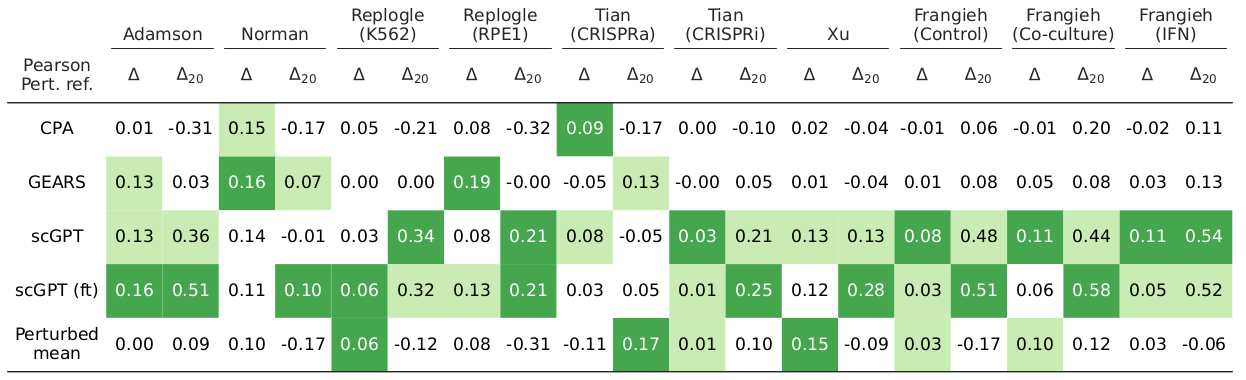
We found that the amount of systematic variation in the perturbation datasets strongly correlated with the performance scores of existing perturbation response prediction methods.
Evaluating the prediction of perturbation-specific effects
To increase robustness to systematic variation, we developed Systema, a new evaluation framework for perturbation response prediction. Instead of using control cells as a point of reference, Systema allows using custom references that better isolate perturbation-specific effects. We then redefined standard evaluation metrics using the perturbed centroid as reference. Application of Systema resulted in substantially lower evaluation scores.
Dissecting the predictable perturbations of perturbation response prediction methods
Existing perturbation response methods struggle to infer the perturbation-specific effects of unseen genetic perturbations. Can these methods still produce biologically informative predictions? To what extent can they uncover coarse-grained effects? To investigate this, we introduce an intuitive evaluation metric in the Systema framework that we refer to as centroid accuracy, which measures whether predicted post-perturbation profiles are closer to their correct ground-truth centroid than to the centroids of other perturbations. A centroid accuracy of 1 indicates that the inferred profiles recover the expected transcriptional effects of a perturbation. We applied this metric to evaluate predictions on unseen 1-gene perturbations across ten datasets and found that the average perturbation scores barely exceeded those of the perturbed mean.
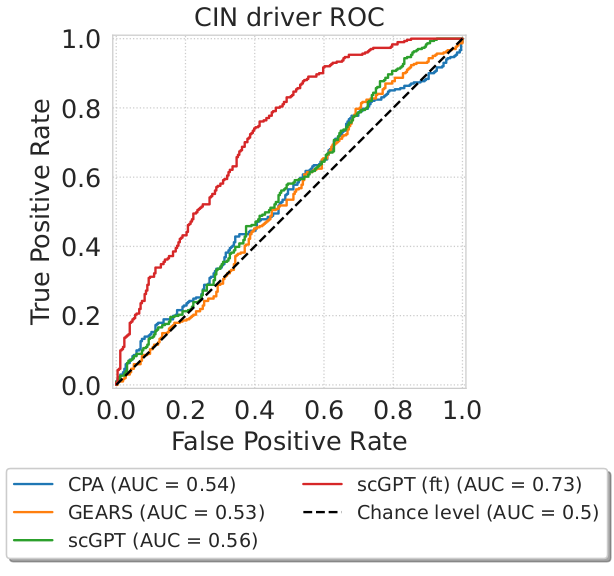
To further evaluate the biological utility of perturbation response predictions, we extended the centroid accuracy to test whether the predicted centroids could distinguish coarse-grained perturbation effects. Specifically, we used the inferred centroids to classify unseen perturbations as inducing either low or high chromosomal instability (CIN) in the genome-wide K562 perturbation screen, using annotations from Replogle et al. (2022). We classified perturbations based on the distances between their inferred centroids and two class-specific centroids, representing perturbations inducing low and high chromosomal instabilities. Among all methods, only the finetuned version of scGPT achieved a ROC-AUC substantially above chance (AUC=0.7).
Conclusion
In this paper, we evaluated the capabilities of existing perturbation response prediction methods and found that their high predictive scores are largely driven by systematic biases. Moreover, we observed that systematic variation can profoundly affect downstream tasks and that existing reference-based metrics are susceptible to systematic effects, which may impair the accurate assessment of perturbation response prediction methods. To isolate perturbation-specific effects, we introduced Systema, a flexible evaluation framework that enables alternative points of reference, including the centroid of perturbed cells. Using this strategy, we observed substantially lower evaluation scores, demonstrating that generalizing to unseen genetic perturbations is a notoriously challenging task.
Looking forward, we believe that perturbation response models should be evaluated based on their biological utility, i.e., how can inferred perturbation profiles help us answer downstream queries about relevant cellular phenotypes? Framing evaluation in terms of downstream tasks may offer a more meaningful and practical perspective. In this light, emerging perturbation platforms like optical pooled screens and spatial functional genomics screens, which combine perturbation data with cell morphology, spatial context, and tissue-level features, present particularly rich opportunities. These modalities may offer access to a broad range of cellular and multicellular phenotypes, allowing predicted gene expression profiles to be studied not as the ultimate goal, but as an intermediate step toward understanding the functional impact of perturbations.
Code
An implementation of our benchmark is available on GitHub.
Data
We downloaded and processed data using the GEARS (Roohani et al., 2023) codebase. The Gene Expression Omnibus accession numbers used are: Adamson et al. (2016): GSE90546, Norman et al. (2019): GSE146194, Xu et al. (2024): GSE218566. The data from Replogle et al. (2022) are available at: https://doi.org/10.25452/figshare.plus.20022944 and additional annotations are available at: https://doi.org/10.25452/figshare.plus.21632564. The Frangieh et al. (2021) data are available at: https://singlecell.broadinstitute.org/single_cell/study/SCP1064/multi-modal-pooled-perturb-cite-seq-screens-in-patient-models-define-novel-mechanisms-of-cancer-immune-evasion. The Tian et al. (2019) data are available via scPerturb (Peidli et al., 2024) at: https://doi.org/10.5281/zenodo.13350497.
Contributors
The following people contributed to this work:
Ramon Viñas Torné
Maciej Wiatrak
Zoe Piran
Shuyang Fan
Liangze Jiang
Sarah A. Teichmann
Mor Nitzan